[paper review] CLUSTER AND SEPARATE: A GNN APPROACH TO VOICE AND STAFF PREDICTION FOR SCORE ENGRAVING
Paper Review
from the paper.
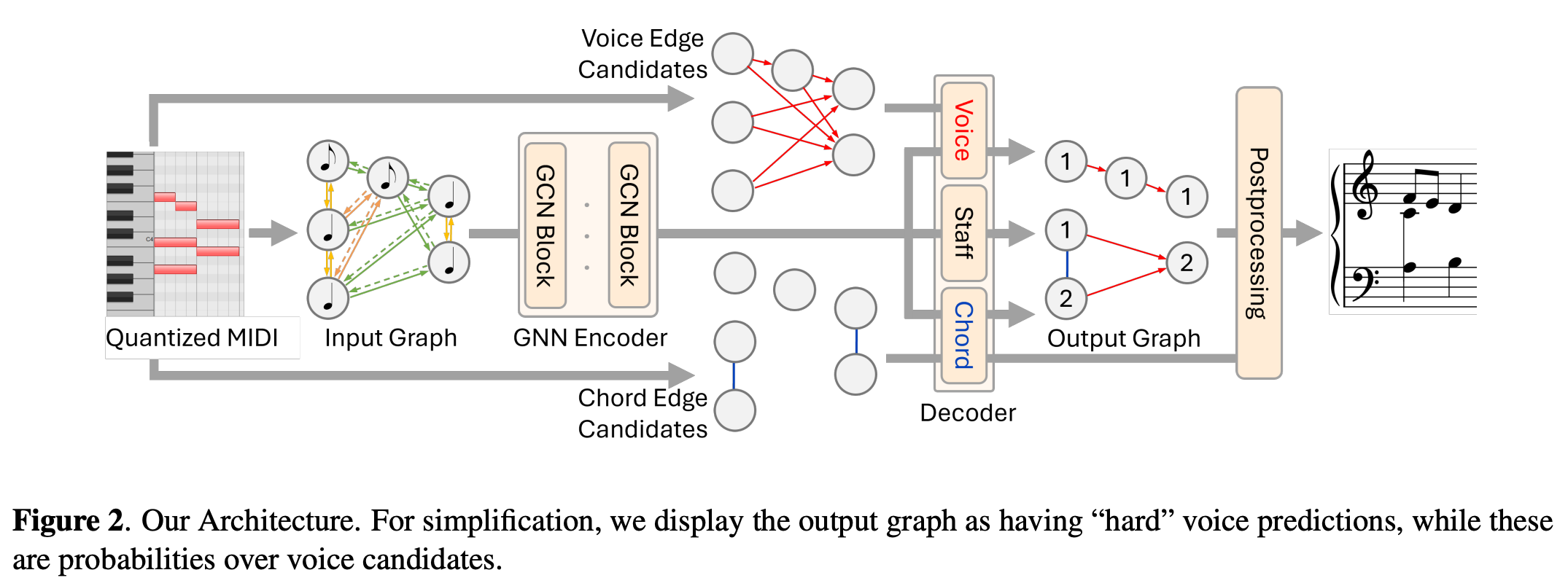
In this post, I would like to review the paper "CLUSTER AND SEPARATE: A GNN APPROACH TO VOICE AND STAFF PREDICTION FOR SCORE ENGRAVING" by Foscarin et al. (2024) presented in the ISMIR2024. I would focus on explaining the overall process by explaining the codes.
Main Predict Function
Load checkpoint & Parsing
It uses Partitura which reads musicxml format based on lxml parsing. AS musicxml format itself contains division information by quarter note, we can't change the value according to our purpose like converting the information into event-based encoding.
# Load the model
pl_model = PLPianoSVSep.load_from_checkpoint(path_to_model, map_location="cpu")
# Prepare the score
pg_graph, score, tied_notes = prepare_score(path_to_score)Example output
# pg_graph
HeteroData(
name='test_graph',
note={
x=[1978, 23],
pitch=[1978],
onset_div=[1978],
duration_div=[1978],
onset_beat=[1978],
duration_beat=[1978],
ts_beats=[1978],
staff=[1978],
voice=[1978],
},
(note, onset, note)={ edge_index=[2, 1416] },
(note, consecutive, note)={ edge_index=[2, 3027] },
(note, during, note)={ edge_index=[2, 802] },
(note, rest, note)={ edge_index=[2, 15] },
(note, potential, note)={ edge_index=[2, 17995] },
(note, truth, note)={ edge_index=[2, 1887] },
(note, chord_potential, note)={ edge_index=[2, 676] },
(note, chord_truth, note)={ edge_index=[2, 350] }
)Prediction step
# Batch for compatibility
pg_graph = pyg.data.Batch.from_data_list([pg_graph])
# predict the voice assignment
with torch.no_grad():
pl_model.module.eval()
pred_voices, pred_staff, pg_graph = pl_model.predict_step(pg_graph, return_graph=True)forward function
if self.rev_edges is not None:
add_reverse_edges(graph, mode=self.rev_edges)
edge_index_dict = graph.edge_index_dict
gbatch = torch.zeros(len(graph.x_dict["note"]), dtype=torch.long, device=graph.x_dict["note"].device)
x_dict = graph.x_dict
pot_edges = edge_index_dict.pop(("note", "potential", "note"))
pot_chord_edges = edge_index_dict.pop(("note", "chord_potential", "note"))
durations = graph["note"].duration_div
onset = graph["note"].onset_div
pitches = graph["note"].pitch
onset_beats = graph["note"].onset_beat
duration_beats = graph["note"].duration_beat
ts_beats = graph["note"].ts_beats
na = torch.vstack((onset_beats, onset, durations, duration_beats, pitches)).t()
# create edge attributes
edge_attr_dict = self.create_edge_attr(na, edge_index_dict)
# which potential edges are in the truth edges
edge_pred_mask_logits, staff_pred_logits, features, pooling_mask_logits = self.module(
x_dict, edge_index_dict, pot_edges, pot_chord_edges, gbatch, onset, durations, pitches, onset_beats,
duration_beats, ts_beats,
edge_attr_dict=edge_attr_dict)
edge_pred_mask_prob = torch.sigmoid(edge_pred_mask_logits)
num_nodes = len(graph.x_dict["note"])
# post-processing using coalesce function
new_edge_index, new_edge_probs, unpool_info, reduced_num_nodes = self.ps_pool(
pot_edges, edge_pred_mask_prob, pot_chord_edges, torch.sigmoid(pooling_mask_logits),
batch=gbatch, num_nodes=num_nodes)
# Assignment problem for edge connection (voicing)
post_monophonic_edges = linear_assignment(new_edge_probs, new_edge_index, reduced_num_nodes, threshold=self.threshold)
post_pred_edges = self.ps_pool.unpool(post_monophonic_edges, reduced_num_nodes, unpool_info)
# return
return_graph = kwargs.get("return_graph", False)
if return_graph:
graph["note", "chord_potential", "note"].edge_index = pot_chord_edges
graph["note", "potential", "note"].edge_index = pot_edges
graph["note", "chord_predicted", "note"].edge_index = pot_chord_edges[:, torch.sigmoid(pooling_mask_logits) > self.threshold]
graph["note", "predicted", "note"].edge_index = post_pred_edges
post_pred_edges = torch.cat(
(post_pred_edges, pot_chord_edges[:, torch.sigmoid(pooling_mask_logits) > self.threshold]), dim=1)
return post_pred_edges, staff_pred_logits.argmax(dim=1).long(), graph
post_pred_edges = torch.cat(
(post_pred_edges, pot_chord_edges[:, torch.sigmoid(pooling_mask_logits) > self.threshold]), dim=1)
# add the chord edges to the post_pred_edges
return post_pred_edges, staff_pred_logits.argmax(dim=1).long()create_edge_attr
difference between each attributes (position, pitch) are added for graph neural network.
edge_attr_dict = {}
for key, value in edge_index_dict.items():
new_v = na[value[0]] - na[value[1]] # difference between source and target nodes
new_v_pitch = self.pitch_embedding(
torch.remainder(new_v[:, -1], 12).long()) # new_v[:, -1] is the pitch, so we take the remainder of 12
new_v = F.normalize(torch.abs(new_v), dim=0)
edge_attr_dict[key] = torch.cat([new_v, new_v_pitch], dim=-1)
return edge_attr_dictself.module = GNN
GNN predicts staff, voice and edge logits.
z_dict = self.encoder(x_dict, edge_index_dict, edge_attr_dict)
hidden_features = z_dict["note"]
hidden_features = self.after_encoder_frontend(hidden_features)
if self.chord_pooling_mode != "none":
hidden_features, pooling_logits = self.pooling_layer(hidden_features, pot_chord_edges, batch)
else:
# create dummy chord edge score
pooling_logits = torch.zeros_like(pot_chord_edges)
staff_logits = self.staff_clf(hidden_features)
out = self.decoder(hidden_features, pot_edges, onsets, durations, pitches, onset_beat, duration_beat, ts_beats, staff_logits)
return out, staff_logits, hidden_features, pooling_logitspost-processing part
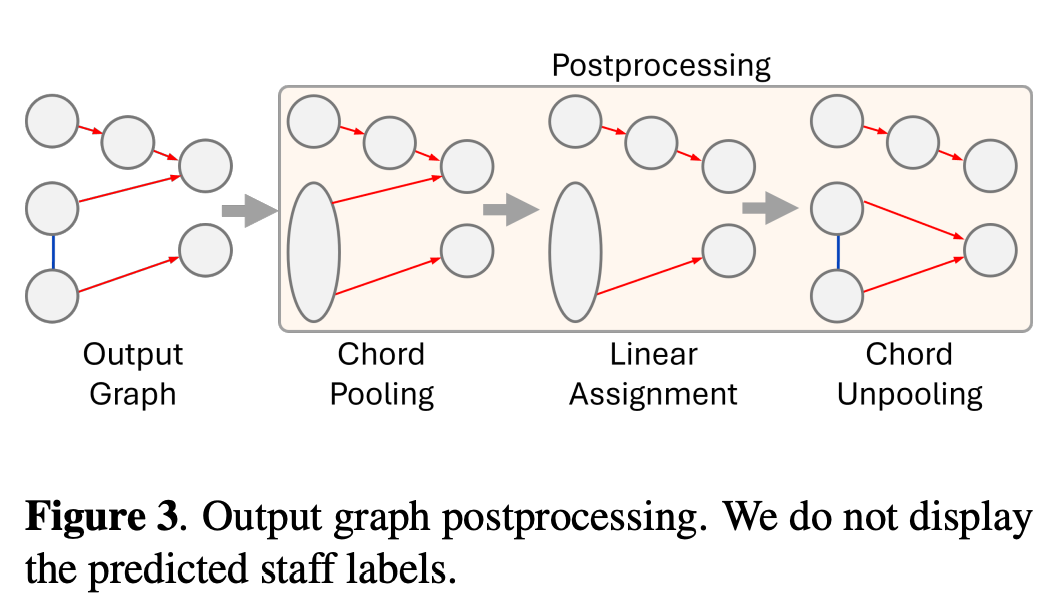
Coalesce is the main function for post-processing. Pytorch supports this function for GNN based work.
from the paper.
What is Colalesce
A Simple Example
Imagine we have a small graph with 4 nodes before pooling, with these original edge indices (each column represents an edge):
edge_index (2 x 5 tensor):
[[0, 0, 1, 2, 2],
[1, 2, 2, 3, 3]]Suppose the cluster mapping (after grouping nodes into chords) is:
cluster = [0, 0, 1, 1]This means:
Nodes 0 and 1 are merged into cluster 0.
Nodes 2 and 3 are merged into cluster 1.
Now, when we remap the edge indices using the cluster mapping:
For each index in edge_index, we replace it with its corresponding value in cluster.
So, after mapping, we get:
new_edge_index_raw =
[[0, 0, 0, 1, 1],
[0, 1, 1, 1, 1]]Let's also assume the corresponding edge_probs (a probability for each edge) are:
edge_probs = [0.8, 0.6, 0.4, 0.9, 0.7]Now, look at the remapped edges:
Edge from 0 to 0: There is one such edge: (0, 0) with probability 0.8.
Edges from 0 to 1: There are two edges:
One from the second edge (0 → 1) with probability 0.6.
Another from the third edge (0 → 1) with probability 0.4.
Edges from 1 to 1: There are two edges:
Both from edges 4 and 5: (1 → 1) with probabilities 0.9 and 0.7.
The coalesce function now merges edges that have the same (source, target) pair:
For the (0, 0) edge: It remains as is, with probability 0.8.
For the (0, 1) edges: They are merged into one edge, and since reduce="mean", the new probability is the average of 0.6 and 0.4, which is:
Mean= (0.6+0.4) / 2 = 0.5
For the (1, 1) edges: They are merged into one edge, with average probability:
Mean= (0.9+0.7) / 2 =0.8
So, the output of coalesce will be:
New edge indices (let’s say sorted in some order):
new_edge_index = [[0, 0, 1],
[0, 1, 1]]Assignment Problem with Hungarian Algorithm

import numpy as np
from scipy.optimize import linear_sum_assignment
# Define the cost matrix (workers x tasks)
cost_matrix = np.array([
[9, 2, 7, 8],
[6, 4, 3, 7],
[5, 8, 1, 8],
[7, 6, 9, 4]
])
# Use the Hungarian algorithm to solve the assignment problem.
# linear_sum_assignment returns two arrays: row indices and column indices.
row_ind, col_ind = linear_sum_assignment(cost_matrix)
# Print the optimal assignment and total cost.
print("Optimal assignment:")
for r, c in zip(row_ind, col_ind):
print(f" Worker {r} assigned to Task {c} (cost: {cost_matrix[r, c]})")
total_cost = cost_matrix[row_ind, col_ind].sum()
print(f"\nTotal minimum cost: {total_cost}")Explanation
-
Cost Matrix Definition:
We create a NumPy array cost_matrix that holds the cost values for each worker-task pair. -
Calling linear_sum_assignment:
The function linear_sum_assignment(cost_matrix) finds the assignment that minimizes the total cost.
It returns two arrays:
row_ind: The indices of the workers.
col_ind: The indices of the tasks assigned to the corresponding worker. -
Printing the Assignment:
We loop over the paired indices from row_ind and col_ind to print which worker is assigned to which task and the corresponding cost. -
Total Cost Calculation:
The total minimum cost is computed by summing the costs of the assigned pairs.
Output
Optimal assignment:
Worker 0 assigned to Task 1 (cost: 2)
Worker 1 assigned to Task 2 (cost: 3)
Worker 2 assigned to Task 0 (cost: 5)
Worker 3 assigned to Task 3 (cost: 4)
Total minimum cost: 14
In short, separating voice is converted into the task as an assignment problem which optimize 1 to 1 connections from all possible connections.
pot_edges.shape
torch.Size([2, 15853])
pot_edges
tensor([[ 0, 0, 0, ..., 1823, 1824, 1824],
[ 120, 121, 122, ..., 1825, 115, 1825]])
new_edge_index.shape
torch.Size([2, 1826])
new_edge_index
tensor([[ 0, 1, 2, ..., 1823, 1824, 1825],
[ 121, 137, 131, ..., 1824, 1825, 1817]])Unpooling
def unpool(self, edge_index, num_nodes, unpool_info: UnpoolInfo) -> Tuple[Tensor, Tensor, Tensor]:
"""
Unpooling operation to reconstruct the original graph from the pooled representation.
Args:
edge_index (Tensor): The edge indices of the pooled graph.
num_nodes (int): The number of nodes in the pooled graph.
unpool_info (UnpoolInfo): The information required for unpooling.
Returns:
Tuple[Tensor, Tensor, Tensor]: The new edge indices after unpooling.
"""
x = torch.arange(num_nodes, device=edge_index.device)
new_x = x[unpool_info.cluster]
new_edge_index = torch.empty((2, 0), dtype=torch.long, device=edge_index.device)
# Reconstruct the edges by creating all possible connections between the nodes in the same cluster
for edge_idx in range(edge_index.shape[-1]):
row = edge_index[0, edge_idx]
col = edge_index[1, edge_idx]
multiple_rows = torch.where(new_x == row)[0]
multiple_cols = torch.where(new_x == col)[0]
new_edges = torch.cartesian_prod(multiple_rows, multiple_cols).T
new_edge_index = torch.cat((new_edge_index, new_edges), dim=-1)
return new_edge_indexExample tensors during the unpooling process
# edge_index.shape -> pooled index
torch.Size([2, 1625])
# edge_index[:, :30]
tensor([[ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 14,
16, 17, 18, 19, 20, 22, 23, 24, 29, 30, 31, 32, 34, 35,
36, 37],
[121, 137, 131, 151, 167, 161, 7, 8, 9, 10, 12, 13, 14, 312,
17, 18, 19, 20, 21, 23, 24, 330, 30, 447, 32, 33, 35, 36,
37, 38]])
# new_x.shape -> record of the clustered nodes
torch.Size([1978])
# new_x[:30]
tensor([116, 0, 0, 0, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126,
127, 128, 129, 130, 1, 1, 2, 2, 2, 2, 131, 132, 133, 134,
135, 136])
# new_edges -> newly constructed nodes
tensor([[1, 2, 3],
[8, 8, 8]])Assign Voices
# predict the voice assignment
with torch.no_grad():
pl_model.module.eval()
pred_voices, pred_staff, pg_graph = pl_model.predict_step(pg_graph, return_graph=True)
# Partitura processing for visualization
part = score[0]
save_path = save_path if save_path is not None else os.path.splitext(path_to_score)[0] + "_pred.mei"
pg_graph.name = os.path.splitext(os.path.basename(save_path))[0]
save_pyg_graph_as_json(pg_graph, ids=part.note_array()["id"], path=os.path.dirname(save_path))
->> assign_voices(part, pred_voices, pred_staff)Adjacency Matrix
predicted_staff = predicted_staff.detach().cpu().numpy().astype(int) + 1 # (make staff start from 1)
note_array = part.note_array()
assert len(part.notes_tied) == len(note_array)
# sort the notes by the note.id to match the order of the note_array["id"] which was used as the input to the model
for i, note in enumerate(part.notes_tied):
note.staff = int(predicted_staff[np.where(note_array["id"] == note.id)[0][0]])
# recompute note_array to include the now newly added staff
note_array = part.note_array(include_staff=True)
preds = predicted_voice_edges.detach().cpu().numpy()
# build the adjacency matrix
graph = sp.sparse.csr_matrix((np.ones(preds.shape[1]), (preds[0], preds[1])), shape=(len(note_array), len(note_array)))
->> n_components, voice_assignment = sp.sparse.csgraph.connected_components(graph, directed=True, return_labels=True)
voice_assignment = voice_assignment.astype(int)# voice_assignment
array([ 0, 1, 1, ..., 200, 200, 200])
# voice_assignment[:30]
array([0, 1, 1, 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 3, 3,
3, 3, 3, 3, 3, 3, 3, 3])Example of using adjacency matrix
Example Graph
Suppose you have 5 nodes (labeled 0 through 4) and the following edges (using a simplified, undirected view for connectivity):
- Edge between node 0 and node 1.
- Edge between node 1 and node 2.
- Edge between node 3 and node 4.
You can represent this as an adjacency matrix:
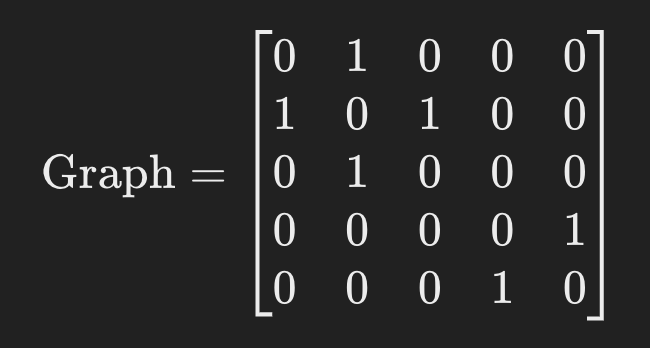
- Interpretation:
Nodes 0, 1, 2 are connected together (forming one group).
Nodes 3, 4 are connected together (forming a second group).
By calling
n_components, voice_assignment = sp.sparse.csgraph.connected_components(graph, directed=True, return_labels=True)You might get:
n_components = 2 (because there are two groups)
voice_assignment = [0, 0, 0, 1, 1]
This means:
Nodes 0, 1, and 2 are all in component 0.
Nodes 3 and 4 are in component 1.