원본데이터 출처 : kaggle
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split, KFold, cross_val_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
from datetime import datetime
import warnings
%matplotlib inline
def encode_features(dataDF):
features = ['Gender','Tumour_Stage','Histology']
for feature in features:
le = LabelEncoder()
le = le.fit(dataDF[feature])
dataDF[feature] = le.transform(dataDF[feature])
return dataDF
def calc (date_str1, date_str2):
date_format = "%d-%b-%y"
date1 = datetime.strptime(date_str1, date_format)
date2 = datetime.strptime(date_str2, date_format)
delta = date2 - date1
return delta.days
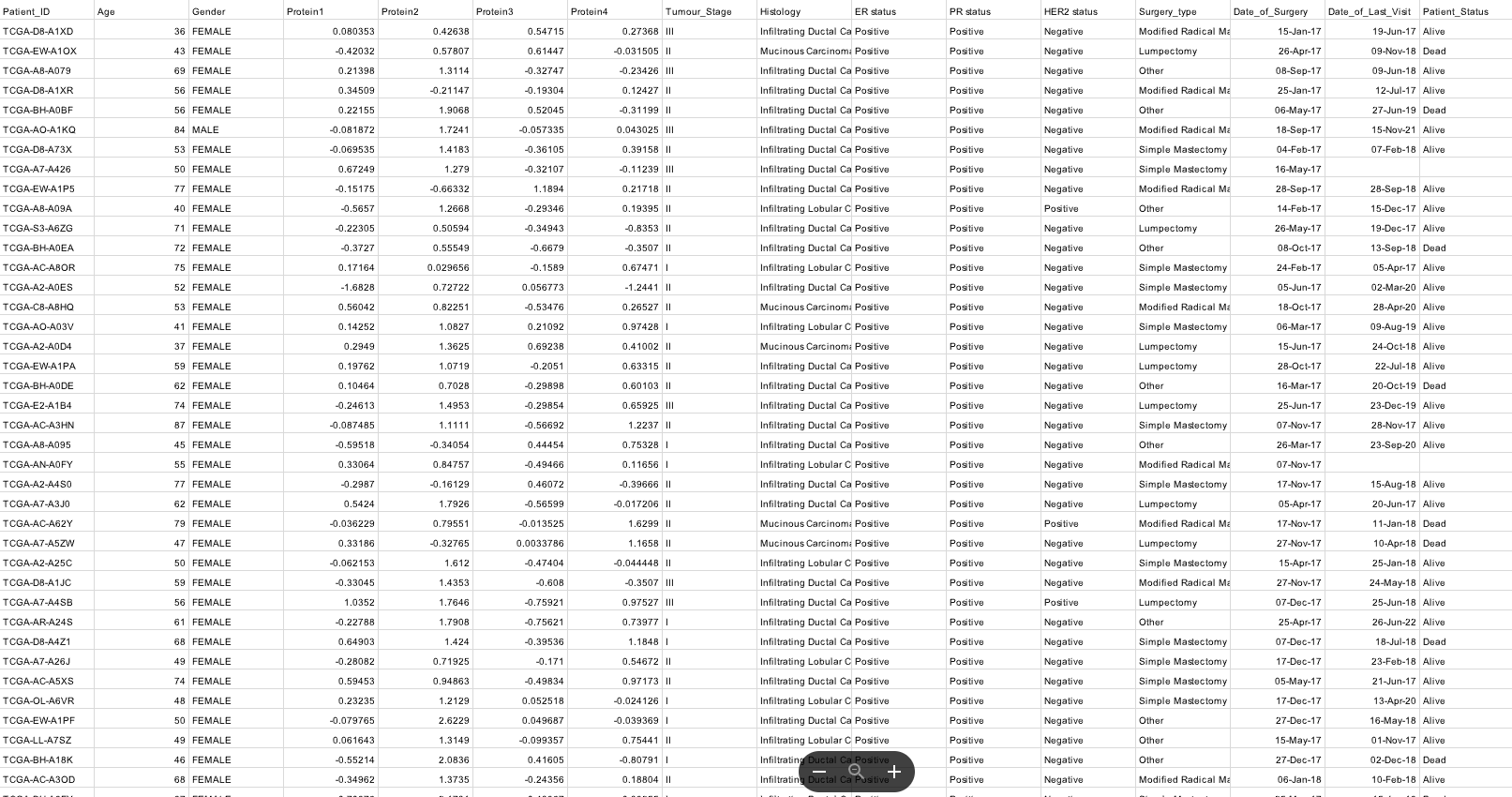
BRCA_df = pd.read_csv(r"C:\Users\User\Data_Handling\BRCA.csv").head(333)
#경고무시
warnings.simplefilter(action='ignore', category=FutureWarning)
#데이터 전처리
BRCA_df['Date_of_Surgery'] = pd.to_datetime(BRCA_df['Date_of_Surgery'], format='%d-%b-%y')
BRCA_df['Date_of_Last_Visit'] = pd.to_datetime(BRCA_df['Date_of_Last_Visit'],format='%d-%b-%y')
BRCA_df['Surgery_Visit_Between'] = BRCA_df['Date_of_Last_Visit'] - BRCA_df['Date_of_Surgery']
BRCA_df = BRCA_df[BRCA_df['Gender']!="MALE"]
BRCA_df = BRCA_df.dropna()
encode_features(BRCA_df)
BRCA_df = BRCA_df.drop(['Patient_ID','Surgery_type','Date_of_Last_Visit','Date_of_Surgery','Gender'],axis=1)
BRCA_df[["ER status","PR status","HER2 status"]] = 1 if "Positive" else 0
BRCA_df["Surgery_Visit_Between"].fillna(BRCA_df["Surgery_Visit_Between"].mean(),inplace=True)
BRCA_df["Surgery_Visit_Between"] = BRCA_df["Surgery_Visit_Between"].apply(lambda x : int(str(x).split()[0]))
#데이터 분류
Y_BRCA_df = BRCA_df["Patient_Status"]
X_BRCA_df = BRCA_df.drop(["Patient_Status"],axis=1)
#머신러닝
dt_clf = DecisionTreeClassifier()
rf_clf = RandomForestClassifier()
lr_clf = LogisticRegression(solver='liblinear')
X_train,X_test,Y_train,Y_test = train_test_split(X_BRCA_df,Y_BRCA_df,test_size=0.2,random_state=11)
#의사결정트리
print("👨🏻⚕️의사결정트리")
dt_clf.fit(X_train,Y_train)
dt_pred = dt_clf.predict(X_test)
print("정확도:",accuracy_score(Y_test,dt_pred))
scores = cross_val_score(dt_clf,X_BRCA_df,Y_BRCA_df,cv=5)
print("Starified_KFold 교차검증 정확도:",scores.mean())
print()
#랜덤포레스트
print("🌳랜덤포레스트")
rf_clf.fit(X_train,Y_train)
rf_pred = rf_clf.predict(X_test)
print("정확도:",accuracy_score(Y_test,rf_pred))
scores = cross_val_score(rf_clf,X_BRCA_df,Y_BRCA_df,cv=5)
print("Starified_KFold 교차검증 정확도:",scores.mean())
print()
#로지스틱회귀
print("💻로지스틱회귀")
lr_clf.fit(X_train,Y_train)
lr_pred = lr_clf.predict(X_test)
scores = cross_val_score(dt_clf,X_BRCA_df,Y_BRCA_df,cv=5)
print("Starified_KFold 교차검증 정확도:",scores.mean())
print("정확도:",accuracy_score(Y_test,lr_pred))
print()



