Overview
Dacon 주최 Computer Vision 이상치 탐지 알고리즘 경진대회에서 MVTec Dataset를 대상으로 사물의 종류와 상태를 분류하는 task를 공부함.
공업에서, image Data보다 Sequence format의 Data를 더 많이 다룰 것으로 판단.
이에 RNN기반 1-step, multi-step Sequence 훈련/예측, 이 Data의 mean, covariance 기반 이상치 탐지를 수행하는 github code를 공부.
본 코드 리뷰는 github of chickenbestlover를 기반.
ECGs 데이터 하나를 타겟으로 LSTM모델 학습.
- Data Format
| Time Step | feature_1 | feature_2 |
|---|---|---|
| 45590.000 | -1.965 | 0.075 |
| 45590.004 | -1.960 | -0.130 |
| 45590.008 | -1.830 | -0.470 |
| 45590.012 | -1.645 | -0.855 |
| 45590.016 | -1.420 | -1.210 |
.....
RNN Code
jupyter notebook에서 argparse를 사용 가능한 형태로 만들기 위해서 easydict.EasyDict 사용.
args = easydict.EasyDict({
'data': 'ecg',
'filename': 'chfdb_chf13_45590.pkl',
'model': 'LSTM',
'augment': True,
'emsize': 32,
'nhid': 32,
'nlayers': 2,
'res_connection': 'store_true',
'lr': 0.0002,
'weight_decay': 1e-4,
'clip': 10,
'epochs': 400,
'batch_size': 64,
'bptt': 50,
'dropout': 0.2,
'device': 'cuda',
'eval_batch_size':64,
'resume': 'False',
'tied': 'store_true',
'pretrained': 'False'
})1. Download dataset with pickle
pickle: txt 파일을 객체처럼 불러오기 위해서 객체 자체를 파일로 저장하는 것.
path.suffix: 파일의 확장자
shutil.unpack_archive: 자동으로 파일 형식을 인식해서 저장소를 만듦.
원본 소스에는 다양한 유형의 데이터가 존재.
아래는 이들 중 1개의 ecg data를 target으로 저장소에 저장하기 위한 코드.
urls = dict()
urls['ecg']=['http://www.cs.ucr.edu/~eamonn/discords/ECG_data.zip']
for dataname in urls:
raw_dir = Path('dataset', dataname, 'raw')
# dataset/dataname/raw 이름의 dir 생성
raw_dir.mkdir(parents=True, exist_ok=True)
for url in urls[dataname]:
filename= raw_dir.joinpath(Path(url).name)
#dataset/dataname/raw/ 경로 아래에 filename
print('Downloading', url)
resp =requests.get(url)
filename.write_bytes(resp.content)
if filename.suffix=='':
filename.rename(filename.with_suffix('.txt'))
#파일에 확장자가 없으면 txt로
print('Saving to', filename.with_suffix('.txt'))
if filename.suffix=='.zip':
print('Extracting to', filename)
unpack_archive(str(filename), extract_dir=str(raw_dir))
#파일 확장자가 zip이면 해당 경로에 압축해제
for filepath in raw_dir.glob('*.txt'):
with open(str(filepath)) as f:
# Data에 이상치 값 labeling as 1
labeled_data=[]
#labeled data를 다른 저장소에 따로 로드할 예정
for i, line in enumerate(f):
tokens = [float(token) for token in line.split()]
if raw_dir.parent.name== 'ecg':
tokens.pop(0)
# ecg data의 경우, time step column이 존재해 1번째 열을 삭제
#해당 dataset의 이상치 정보를 기반으로 이상치 sequence에 label
if filepath.name == 'chfdb_chf13_45590.txt':
tokens.append(1.0) if 2800 < i < 2960 else tokens.append(0.0)
labeled_data.append(tokens)
#anomaly points가 포함된 dataset
#tokens = [[value, 0or1]]
#tokens가 1인 곳이 이상치 0인 곳이 정상치
# labeled data, dataset/dataname/labeled/whole 경로에 pkl(pickle) 형식으로 저장
labeled_whole_dir = raw_dir.parent.joinpath('labeled', 'whole')
labeled_whole_dir.mkdir(parents=True, exist_ok=True)
with open(str(labeled_whole_dir.joinpath(filepath.name).with_suffix('.pkl')), 'wb') as pkl:
pickle.dump(labeled_data, pkl)
# train/test set으로 나눠서 dataset/ecg/labeled/train or test에 pkl 형식으로 저장
# testset이 train 다음 sequence인 모양
labeled_train_dir = raw_dir.parent.joinpath('labeled','train')
labeled_train_dir.mkdir(parents=True,exist_ok=True)
labeled_test_dir = raw_dir.parent.joinpath('labeled','test')
labeled_test_dir.mkdir(parents=True,exist_ok=True)
if filepath.name == 'chfdb_chf13_45590.txt':
with open(str(labeled_train_dir.joinpath(filepath.name).with_suffix('.pkl')), 'wb') as pkl:
pickle.dump(labeled_data[:2439], pkl)
with open(str(labeled_test_dir.joinpath(filepath.name).with_suffix('.pkl')), 'wb') as pkl:
pickle.dump(labeled_data[2439:3726], pkl)2. Data Load & Preprocess
open(path, mode)
file 읽고 쓰는 함수.
non-text file(binary file)는 'rb'로 읽고, 'wb'로 씀.
Tensor.expand_as(other)
Tensor를 other.size 크기로 확장해 반환.
torch.narrow(dim, start, length)
dim: 축소할 차원, start: length를 셀 차원, length: start에서 length 만큼 떨어진 차원 축소.
torch.view(shape)
입력받은 shape으로 tensor 변환.
torch.transpose(dim0, dim1)
dim0, dim1를 전환한 tensor 반환.
torch.contiguous()
contiguous 참조
tensor manipulation(view, narrow, expand, transpose, permute 등) 이후, 축단위 포인터 연산시 contiguous 사용이 필요.
def normalization(seqData, max, min):
return (seqData-min)/(max-min)
def standardization(seqData, mean, std):
return(seqData-mean)/std
# standardization의 역순
def reconstruct(seqData, mean, std):
return seqData*std + mean
class PickleDataLoader(object):
def __init__(self, data_type, filename, augment_test_data = True):
self.augment_test_data=augment_test_data
self.trainData, self.trainLabel = self.preprocessing(Path('dataset',
data_type, 'labeled','train', filename), train=True)
self.testData , self.testLabel = self.preprocessing(Path('dataset',
data_type, 'labeled','test', filename), train=False)
def augmentation(self, data, label, noise_ratio=0.05, noise_interval=0.005, max_length=100000):
# noise 더한 data 행 붙이기(dim=0 => 행 생성해서 붙임)
noiseSeq = torch.randn(data.size())
augmentedData = data.clone()
augmentedLabel = label.clone()
for i in np.arange(0, noise_ratio, noise_interval):
scaled_noiseSeq = noise_ratio*self.std.expand_as(data)*noiseSeq
augmentedData = torch.cat([augmentedData, data + scaled_noiseSeq], dim=0)
augmentedLabel = torch.cat([augmentedLabel, label])
if len(augmentedData) > max_length:
augmentedData = augmentedData[:max_length]
augmentedLabel = augmentedLabel[:max_length]
break
return augmentedData,augmentedLabel
def preprocessing(self, path, train=True):
# data pickle load해서 label이랑 data 떼어놓고
# train mode면 mean std length 구하고, augmentation
# test mode면 augmentation이나 하고
# train에서 구한 mean std length로 train이건 test이건 데이터 정규화
# data, label 출력
with open(str(path), 'rb') as f:
data= torch.FloatTensor(pickle.load(f))
label = data[:,-1]
data = data[:,:-1]
if train:
self.mean = data.mean(dim=0)
self.std = data.std(dim=0)
self.length = len(data)
data, label = self.augmentation(data, label)
else:
if self.augment_test_data:
data, label =self.augmentation(data, label)
data = standardization(data, self.mean, self.std)
return data, label
def batchify(self, args, data, bsz):
# batch를 나누는데 나누어 떨어지지 않는 행 삭제
# narrow 사용으로 column 기준으로 연산 축이 바뀌어 contiguous 처리
nbatch = data.size(0) // bsz
#[len(data),2]
trimmed_data =data.narrow(0,0,nbatch*bsz)
# batch_size로 나누어 떨어지지 않는 열 제거
# size: (n_batch*bsz,2)
batched_data = trimmed_data.contiguous().view(bsz, -1, trimmed_data.size(-1)).transpose(0,1) # size: (nbatch,bsz,2)
batched_data = batched_data.to(device(args.device))
return batched_data3. Model Compose
torch.nn.Module.load_state_dict: model, optimisze의 parameters를 저장하는 python dictionary
getattr(object, 'name'): python 내장함수, object.name과 같음. 다만 name에 string를 attribute할 수 있다는 것이 유용.
shutil: 파일에 대한 고수준 연산 제공.
shutil.copyfile(src, dst): src 파일 내용을 dst 파일에 복사 후 dst 반환.
class RNNPredictor(nn.Module):
def __init__(self, rnn_type, enc_inp_size, rnn_inp_size, rnn_hid_size,
dec_out_size, nlayers, dropout=0.5, tie_weights=False, res_connection=False):
super(RNNPredictor, self).__init__()
self.enc_input_size = enc_inp_size
# feature_dim
self.drop = nn.Dropout(dropout)
self.encoder = nn.Linear(enc_inp_size, rnn_inp_size)
if rnn_type in ['LSTM','GRU']:
self.rnn = getattr(nn, rnn_type)(rnn_inp_size, rnn_hid_size, nlayers, dropout=dropout)
# rnn_type를 str로 받아서 attribute 가능
else:
try:
nonlinearity = {'RNN_TANH': 'tanh', 'RNN_RELU': 'relu'}[rnn_type]
except KeyError:
raise ValueError('''An invalid option for '--model' was supplied,
options are ['LSTM', 'GRU', 'SRU', 'RNN_TANH' or 'RNN_RELU']''')
self.rnn = nn.RNN(rnn_inp_size, rnn_hid_size, nlayers, nonlinearity=nonlinearity, dropout=dropout)
self.decoder = nn.Linear(rnn_hid_size, dec_out_size)
# decoder에 encoder의 가중치를 transpose해서 갖다 쓰는 것.
if tie_weights:
if rnn_hid_size != rnn_inp_size:
raise ValueError('When using the tied flag, nhid must be equal to emsize')
self.decoder.weight = self.encoder.weight
self.res_connection = res_connection
self.rnn_type = rnn_type
self.rnn_hid_size = rnn_hid_size
self.nlayers = nlayers
def init_weights(self):
initrange = 0.1
self.encoder.weight.data.uniform_(-initrange, initrange)
self.decoder.bias.data.fill_(0)
self.decoder.weight.data.uniform_(-initrange, initrange)
def forward(self, input, hidden, return_hiddens=False, noise=False):
emb = self.drop(self.encoder(input.contiguous().view(-1,self.enc_input_size)))
# tensor(64,2) => (64,32)
emb = emb.view(-1, input.size(1), self.rnn_hid_size)
# (1,64,32)
if noise:
hidden = (F.dropout(hidden[0], training=True,p=0.9),F.dropout(hidden[1],training=True,p=0.9))
# double hidden_layer
output, hidden = self.rnn(emb, hidden)
output = self.drop(output)
decoded = self.decoder(output.view(output.size(0)*output.size(1), output.size(2)))
decoded = decoded.view(output.size(0), output.size(1), decoded.size(1))
if self.res_connection:
decoded = decoded + input
if return_hiddens:
return decoded,hidden,output
return decoded, hidden
def init_hidden(self, bsz):
weight = next(self.parameters()).data
if self.rnn_type == 'LSTM':
return(Variable(weight.new(self.nlayers, bsz, self.rnn_hid_size).zero_()),
Variable(weight.new(self.nlayers, bsz, self.rnn_hid_size).zero_()))
else:
return Variable(weight.new(self.nlayers, bsz, self.rnn_hid_size).zero_())
def repackage_hidden(self, h):
if type(h) == tuple:
return tuple(self.repackage_hidden(v) for v in h)
else:
return h.detach()
def save_checkpoint(self, state, is_best):
print('=> saving_checkpoint')
args = state['args']
checkpoint_dir = Path('save',args.data,'checkpoint')
checkpoint_dir.mkdir(parents=True, exist_ok=True)
checkpoint = checkpoint_dir.joinpath(args.filename).with_shffix('.pth')
# save/dataname/checkpoint/filename.pth
torch.save(state, checkpoint)
if is_best:
model_best_dir = Path('save', args.data, 'model_best')
model_best_dir.mkdir(parents=True, exist_ok=True)
# save/dataname/model_best
shutil.copyfile(checkpoint, model_best_dir.joinpath(args.filename).with_suffix('.pth'))
# file를 model_best_dir에 복사
print('=> checkpoint saved.')
def extract_hidden(self, hidden):
if self.rnn_type == 'LSTM':
return hidden[0][-1].data.cpu()
else:
return hidden[-1].data.cpu()
def initialize(self, args, featur_dim):
self.__init__(rnn_type = args.model,
enc_inp_size = feature_dim,
c = args.emsize,
rnn_hid_size = args.nhid,
dec_out_size = feature_dim,
nlayers = args.nlayers,
dropout = args.dropout,
tie_weights = args.tied,
res_connection = args.res_connection)
self.to(args.device)
def load_checkpoint(self, args, checkpoint, feakture_dim):
start_epoch = checkpoint['epoch']+1
best_val_loss = checkpoint['best_loss']
args_ = checkpoint['args']
args_.resume = args.resume
args_.pretrained = args.pretrained
args_.epochs = args.epochs
args_.save_interval = args.save_interval
args_.prediction_window_size = args.prediction_window_size
self.initialize(args_, feature_dim=feature_dim)
self.load_state_dict(checkpoint['state_dict'])
return args_, start_epoch, best_val_loss
def generate_output(args, epoch, model, gen_dataset, disp_uncertainty=True, startPoint=500, endPoint=3500):
if args.save_fig:
model.eval()
hidden = model.init_hidden(1)
# hidden state 가중치 초기화 zero 행렬(default: 2,1,32)
#LSTM 경우, ((2,1,32),(2,1,32))
outSeq=[]
with torch.no_grad():
for i in range(endPoint):
if i >= startPoint:
# 아래서 구한 startPoint value input으로 endPoint value를 출력
out, hidden = model.forward(out, hidden)
# forward(input, hidden)
# input seq에서 embedding: nn.Linear(enc_inp_size, rnn_inp_size) & nn.Dropout(dropout) & reshape
# embedding에서 output, hidden: nn.rnn(emb, hidden)
# output decode: nn.Dropout & nn.Linear(hid_size, out_size) & reshape
# decoded, hidden 반환
outSeq.append(out.data.cpu()[0][0].unsqueeze(0))
else:
# from i=0 to i=startPoint-1
# gen_dataset => batch size 1의 (testdata_len, 1, feature_dim)
# gen_dataset[i].unsqueeze(0) => (1,1,feature_dim), hidden => (2,1,32)
out, hidden = model.forward(gen_dataset[i].unsqueeze(0),hidden)
# startpoint index까지 찾아가는 과정
outSeq.append(out.data.cpu()[0][0].unsqueeze(0))
outSeq = torch.cat(outSeq, dim=0)
target = reconstruct(gen_dataset.cpu(),TimeseriesData.mean, TimeseriesData.std)
outSeq = reconstruct(outSeq, TimeseriesData.mean, TimeseriesData.std)
# Data*std + mean
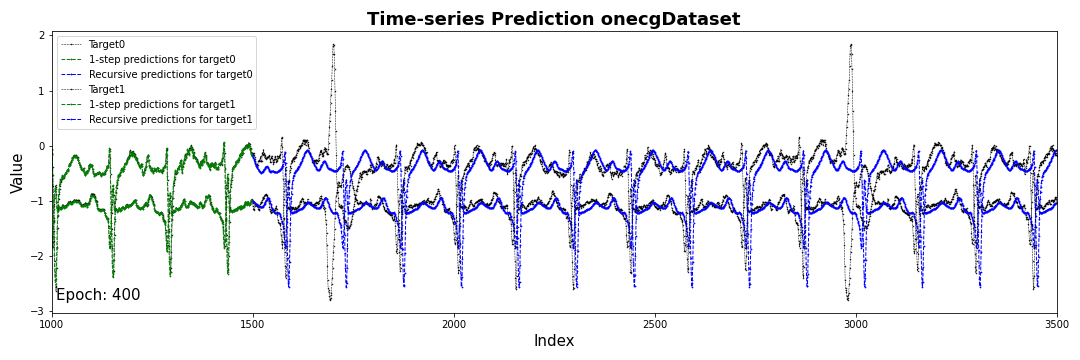
plt.figure(figsize=(15,5))
for i in range(target.size(-1)):
plt.plot(target[:,:,i].numpy(), label='Target'+str(i),
color='black', marker='.', linestyle='--', markersize=1, linewidth=0.5)
plt.plot(range(startPoint), outSeq[:startPoint,i].numpy(), label='1-step predictions for target'+str(i),
color ='green', marker='.', linestyle = '--', markersize=1.5, linewidth=1)
plt.plot(range(startPoint, endPoint), outSeq[startPoint:,i].numpy(), label='Recursive predictions for target'+str(i),
color='blue', marker='.',linestyle='--', markersize=1.5, linewidth=1)
plt.xlim([startPoint-500, endPoint])
plt.xlabel('Index',fontsize=15)
plt.ylabel('Value',fontsize=15)
plt.title('Time-series Prediction on' + args.data + 'Dataset', fontsize=18, fontweight='bold')
plt.legend()
plt.tight_layout()
plt.text(startPoint-500+10, target.min(), 'Epoch: ' + str(epoch), fontsize=15)
save_dir = Path('result', args.data, args.filename).with_shffix('').joinpath('fig_prediction')
save_dir.mkdir(parents=True, exist_ok=True)
plt.savefig(save_dir.joinpath('fig_epoch'+str(epoch)).with_shffix('.png'))
plt.close()
return outSeq
else:
pass4. Sequence training/evaluate Execute
TimeseriesData = PickleDataLoader(data_type=args.data, filename=args.filename,
augment_test_data=args.augment)
train_dataset = TimeseriesData.batchify(args, TimeseriesData.trainData, args.batch_size)
test_dataset = TimeseriesData.batchify(args, TimeseriesData.testData, args.eval_batch_size)
gen_dataset = TimeseriesData.batchify(args, TimeseriesData.testData, 1)
feature_dim = TimeseriesData.trainData.size(1) #2
model = RNNPredictor(rnn_type = args.model,
enc_inp_size = feature_dim,
rnn_inp_size = args.emsize,
rnn_hid_size = args.nhid,
dec_out_size = feature_dim,
nlayers = args.nlayers,
dropout = args.dropout,
tie_weights = args.tied,
res_connection = args.res_connection
).to(args.device)
optimizer = optim.Adam(model.parameters(), lr=args.lr, weight_decay=args.weight_decay)
criterion = nn.MSELoss()#bptt값 만큼 seq를 떼어서 batch 구성
#target은 data의 한칸 앞 seq
def get_batch(args,source, i):
seq_len = min(args.bptt, len(source) -1 -i)
data = source[i:i+seq_len]
target = source[i+1:i+1+seq_len]
return data, target
def train(args, model, train_dataset, epoch):
with torch.enable_grad():
model.train()
total_loss=0
start_time = time.time()
hidden = model.init_hidden(args.batch_size) # zeros(layer_dim, batch_size, hidden_size) hidden state 초기화
## 여기서부터 1-step train
for batch, i in enumerate(range(0, train_dataset.size(0)-1, args.bptt)):
inputSeq, targetSeq = get_batch(args, train_dataset, i) # bptt만틈 떼어내서 batch를 구성, 해당 bptt항 바로 한칸 뒤의 sequence를 targetSeq로 지정
hidden = model.repackage_hidden(hidden)
hidden_ = model.repackage_hidden(hidden)
optimizer.zero_grad()
outVal = inputSeq[0].unsqueeze(0) # (1,64,2), bptt개의 sequence 중 첫번째 항
outVals = []
hids1 = []
for i in range(inputSeq.size(0)):
outVal, hidden_, hid = model.forward(outVal, hidden_, return_hiddens=True) # input(1,64,2)/ emb(1,64,32)/ hidden(2,64,32)/ output(1,64,2)
outVals.append(outVal) # output 하나씩 학습해서 bptt개의 예측값 outVals에 저장
hids1.append(hid)
outSeq1 = torch.cat(outVals, dim=0)# outSeq1.size => (50,64,2), 하나의 행으로
hids1 = torch.cat(hids1, dim=0)
loss1 = criterion(outSeq1.contiguous().view(args.batch_size,-1), targetSeq.contiguous().view(args.batch_size,-1))
# targetSeq와 loss 계산
# loss를 계산할 때, 3 dimension (50,64,2) 자체로 연산을 하면 학습이 잘 이루어지지 않아 (64,100)으로 차원을 줄여 연산
## 여기서부터 multi-step train
# machanism 동일
outSeq2, hidden, hids2 = model.forward(inputSeq, hidden, return_hiddens=True)
loss2 = criterion(outSeq2.contiguous().view(args.batch_size,-1), targetSeq.contiguous().view(args.batch_size,-1))
loss3 = criterion(hids1.view(args.batch_size,-1), hids2.view(args.batch_size,-1).detach()) # hidden state 가중치 loss 계산
loss = loss1+loss2+loss3
loss.backward()
optimizer.step()
total_loss += loss.item()
def evaluate(args, model, test_dataset):
# train과 같은 방식으로 test_dataset를 가지고 eval, total loss 반환
model.eval()
with torch.no_grad():
total_loss = 0
hidden = model.init_hidden(args.eval_batch_size)
nbatch = 1
for nbatch, i in enumerate(range(0, test_dataset.size(0)-1, args.bptt)):
inputSeq, targetSeq = get_batch(args, test_dataset, i)
hidden_ = model.repackage_hidden(hidden)
outVal = inputSeq[0].unsqueeze(0)
outVals = []
hids1= []
for i in range(inputSeq.size(0)):
outVal, hidden_, hid = model.forward(outVal, hidden_, return_hiddens=True)
outVals.append(outVal)
hids1.append(hid)
outSeq1 = torch.cat(outVals, dim=0)
hids1 = torch.cat(hids1, dim=0)
loss1 = criterion(outSeq1.contiguous().view(args.batch_size,-1), targetSeq.contiguous().view(args.batch_size,-1))
outSeq2, hidden, hids2 = model.forward(inputSeq, hidden, return_hiddens=True)
loss2 = criterion(outSeq2.contiguous().view(args.batch_size,-1), targetSeq.contiguous().view(args.batch_size,-1))
loss3 = criterion(hids1.view(args.batch_size,-1), hids2.view(args.batch_size,-1).detach())
loss = loss1+loss2+loss3
total_loss += loss.item()
return total_loss/ (nbatch+1)if args.resume =='True' or args.pretrained == 'True':
print('=> loading checkpoint')
checkpoint = torch.load(Path('save', args.data, 'checkpoint', args.filename).with_suffix('.pth'))
args, start_epoch, best_val_loss = model.load_checkpoint(args.checkpoint, feature_dim)
optimizer.load_state_dict((checkpoint['optimizer']))
del checkpoint
epoch = start_epoch
print('=> loaded checkpoint')
else:
epoch=1
start_epoch=1
best_val_loss = float('inf')
print('=> Start training from scratch')
print('-'*89)
print(args)
print('-'*89)
if args.pretrained == 'False':
try:
for epoch in range(start_epoch, args.epochs+1):
epoch_start_time = time.time()
train(args, model, train_dataset, epoch)
val_loss = evaluate(args, model, test_dataset)
print('-'*89)
print('| end of epoch {:3d} | time: {:5.2f}s | valid loss {:5.4f} |'.format(epoch, (time.time-epoch_start_time), val_loss))
print('-'*89)
generate_output(args, epoch, model, gen_dataset, startPoint=1500)
# model 거친 output sequence 반환
if epoch%args.save_interval==0:
#일정 간격마다 val_loss 체크해서 best를 갱신
is_best = val_loss < best_val_loss
best_val_loss = min(val_loss, best_val_loss)
model_dictionary = {'epoch': epoch,
'best_loss': best_val_loss,
'state_dict': model.state_dict(),
'optimizer': optimizer.state_dict(),
'args':args
}
model.save_checkpoint(model_dictionary, is_best)
except KeyboardInterrupt:
print('-'*89)
print('Exiting from training early')RNN Conclusion
*코드 별 설명은 해당 코드 아래 주석으로 설명.
1. 시계열 Dataset train/test 구간 설정
2. Encoder+Rnn+Decoder로 구성된 모델 설정
3. 50 sequence씩 batch 구성
4. 1-step prediction를 통한 loss1, 50seq prediction를 통한 loss2, loss1, loss2 연산에서 학습한 hidden state를 통한 loss3
5. MSELoss/Adam 으로 모델 학습
6. genuine data와 같이 plot
Anomaly Dectection
Overview
- TrainDataset으로 feature별 예측값과 실제값의 편차의 mean, cov 도출.
- TrainDataset, mean. cov를 input으로
Multivariate Gaussian Distribution Model로 score 도출. - score, label을 가지고 정밀도, 재현율, f1 score curve를 바탕으로 타당한 threshold 도출.
- threshold 초과 score는 이상치로 판단.
- Mulltivariate Gaussian Distribution Model
변수 여러개를 다루는 기본적인 확률분포 모델로 변수들의 평균과 공분산을 바탕으로 표준적인 확률분포를 생성함.
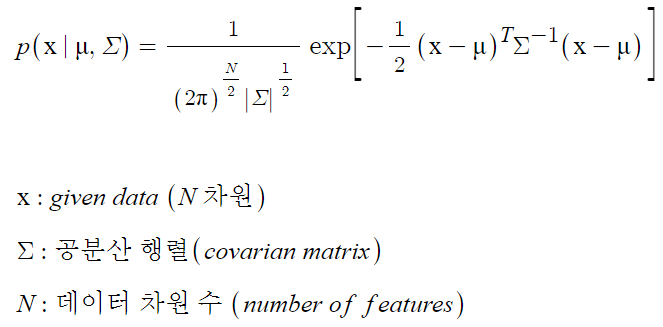
본 코드에서는 확률분포함수의 exp 뒤 부분을 score로 설정해 사용.
-
왜 다변량인가?
=> error_tensor로 지정한 변수에 대한 확률분포함수를 구해 score를 도출하는데, error_tensor의 형태가 다음과 같음.[[a10-10 - mean,a10-9 - mean,a10-8 - mean,...,a10-1 - mean]], [a11-10 - mean,a11-9 - mean,a11-8 - mean,...,a11-1 - mean], ...]
a(n-m)는 예측치, n=시작 지점으로부터 nth value, m=몇 step 전 값으로부터 예측.
nth 편차들이 변수로 작용해 다변량 확률분포를 예측.
학습 데이터의 mean, cov를 이용해 다변량 가우시안 분포를 생성해 각 변수에 대한 anomaly score를 계산하고 임계치를 학습해 이상치를 탐지.
Code
def fit_norm_distribution_param(args, model, train_dataset, channel_idx=0):
# train_dataset value 하나씩 모델에 학습해 예측한 값 한개를 predictions에 넣음
# window_size(n) 만큼 iter동안 predictions에 [[a1,...an],[a2,...an+1],...[an+1...a2n]] format으로 저장
# 같은 iter동안 organized, errors list는 [[]1,[],...,[]n] format 형성
# window_size(n) iter 이후로는 organized에 [[an1,an2,...,ann]] format으로 각 step에서 예측한 n번째 값 n개를 저장
# errors에는 organized값과 실제값과의 편차를 저장
predictions = []
organized = []
errors = []
with torch.no_grad():
model.eval()
pasthidden = model.init_hidden(1)#batch_size
for t in range(len(train_dataset)):
out, hidden = model.forward(train_dataset[t].unsqueeze(0), pasthidden)
predictions.append([])
organized.append([])
errors.append([])
predictions[t].append(out.data.cpu()[0][0][channel_idx])
pasthidden = model.repackage_hidden(hidden)
for prediction_step in range(1, args.prediction_window_size):
out, hidden = model.forward(out, hidden)
predictions[t].append(out.data.cpu()[0][0][channel_idx])
if t >= args.prediction_window_size:
for step in range(args.prediction_window_size):
organized[t].append(predictions[step+t-args.prediction_window_size][args.prediction_window_size-1-step])
organized[t] = torch.FloatTensor(organized[t]).to(args.device)
errors[t] = organized[t]-train_dataset[t][0][channel_idx]
errors[t] = errors[t].unsqueeze(0)
errors_tensor = torch.cat(errors[args.prediction_window_size:],dim=0)
mean = errors_tensor.mean(dim=0)
# mean은 편차를 1x10 으로 평균해놓은 것
cov = errors_tensor.t().mm(errors_tensor)/errors_tensor.size(0) - mean.unsqueeze(1).mm(mean.unsqueeze(0))
# 공분산은 (편차T x 편차)의 평균 - 평균 x 평균T
return mean,covdef anomalyScore(args, model, dataset, mean, cov, channel_idx=0, score_predictor=None):
# train set으로 구한 mean과 cov를 사용해 지표가 될 score를 구함.
# multivariate gaussian distribution model 사용
# score = (편차-평균) x 공분산 inverse x (편차-평균)T
predictions = []
rearranged = []
errors = []
hiddens = []
predicted_scores = []
with torch.no_grad():
model.eval()
pasthidden = model.init_hidden(1)
for t in range(len(dataset)):
out, hidden = model.forward(dataset[t].unsqueeze(0), pasthidden)
predictions.append([])
rearranged.append([])
errors.append([])
hiddens.append(model.extract_hidden(hidden))
if score_predictor is not None:
predicted_scores.append(score_predictor.predict(model.extract_hidden(hidden).numpy()))
predictions[t].append(out.data.cpu()[0][0][channel_idx])
pasthidden = model.repackage_hidden(hidden)
for prediction_step in range(1, args.prediction_window_size):
out, hidden = model.forward(out, hidden)
predictions[t].append(out.data.cpu()[0][0][channel_idx])
if t >= args.prediction_window_size:
for step in range(args.prediction_window_size):
rearranged[t].append(
predictions[step + t - args.prediction_window_size][args.prediction_window_size - 1 - step])
rearranged[t] =torch.FloatTensor(rearranged[t]).to(args.device).unsqueeze(0)
errors[t] = rearranged[t] - dataset[t][0][channel_idx]
else:
rearranged[t] = torch.zeros(1,args.prediction_window_size).to(args.device)
errors[t] = torch.zeros(1, args.prediction_window_size).to(args.device)
predicted_scores = np.array(predicted_scores)
scores = []
for error in errors:
mult1 = error-mean.unsqueeze(0) # [ 1 * prediction_window_size ]
mult2 = torch.inverse(cov) # [ prediction_window_size * prediction_window_size ]
mult3 = mult1.t() # [ prediction_window_size * 1 ]
score = torch.mm(mult1,torch.mm(mult2,mult3))
scores.append(score[0][0])
scores = torch.stack(scores)
# dataset 길이 - 10개의 num
rearranged = torch.cat(rearranged,dim=0)
errors = torch.cat(errors,dim=0)
return scores, rearranged, errors, hiddens, predicted_scoresdef get_precision_recall(args, score, label, num_samples, beta=1.0, sampling='log', predicted_score=None):
'''
:param args:
:param score: anomaly scores
:param label: anomaly labels
:param num_samples: the number of threshold samples
:param beta:
:param scale:
:return:
'''
if predicted_score is not None:
score = score - torch.FloatTensor(predicted_score).squeeze().to(args.device)
maximum = score.max()
if sampling=='log':
# Sample thresholds logarithmically
# The sampled thresholds are logarithmically spaced between: math:`10 ^ {start}` and: math:`10 ^ {end}`.
th = torch.logspace(0, torch.log10(torch.tensor(maximum)), num_samples).to(args.device)
else:
# Sample thresholds equally
# The sampled thresholds are equally spaced points between: attr:`start` and: attr:`end`
th = torch.linspace(0, maximum, num_samples).to(args.device)
precision = []
recall = []
for i in range(len(th)):
# num_samples만큼 threshold 반복 실험
anomaly = (score > th[i]).float()
# score가 임계치 초과이면 1(이상치) nor 0(정상치)
idx = anomaly * 2 + label
## 다음 변수들은 해당하는 항의 갯수
tn = (idx == 0.0).sum().item()
# tn => 정상치로 예측(anomaly=0), 실제 정상치(label=0)
fn = (idx == 1.0).sum().item()
# fn => 정상치로 예측(anomaly=0), 실제 이상치(label=1)
fp = (idx == 2.0).sum().item()
# fp => 이상치로 예측(anomaly=1), 실제 정상치(label=0)
tp = (idx == 3.0).sum().item()
# tp => 이상치로 예측(anomaly=1), 실제 이상치(label=1)
p = tp / (tp + fp + 1e-7)
# 분모 0 방지
# 정밀도(이상치로 예측한 것 중에 정답 비율)
r = tp / (tp + fn + 1e-7)
# 재현율(실제 이상치 중에 이상치로 예측한 비율)
if p != 0 and r != 0:
precision.append(p)
recall.append(r)
precision = torch.FloatTensor(precision)
recall = torch.FloatTensor(recall)
f1 = (1 + beta ** 2) * (precision * recall).div(beta ** 2 * precision + recall + 1e-7)
#f1 score
return precision, recall, f1print('=> calculating mean and covariance')
means, covs = list(), list()
train_dataset = TimeseriesData.batchify(args, TimeseriesData.trainData, bsz=1)
for channel_idx in range(model.enc_input_size):
mean, cov = fit_norm_distribution_param(args, model, train_dataset[:TimeseriesData.length], channel_idx)
means.append(mean), covs.append(cov)
model_dictionary = {'epoch': max(epoch, start_epoch),
'best_loss': best_val_loss,
'state_dict': model.state_dict(),
'optimizer': optimizer.state_dict(),
'args': args,
'means': means,
'covs': covs
}
model.save_checkpoint(model_dictionary, True)
print('-'*89)TimeseriesData = PickleDataLoader(data_type=args.data, filename=args.filename, augment_test_data=False)
train_dataset = TimeseriesData.batchify(args,TimeseriesData.trainData[:TimeseriesData.length],bsz=1)
test_dataset = TimeseriesData.batchify(args,TimeseriesData.testData,bsz=1)
n_features = TimeseriesData.trainData.size(-1)
model = RNNPredictor(rnn_type = args.model,
enc_inp_size = n_features,
rnn_inp_size = args.emsize,
rnn_hid_size = args.nhid,
dec_out_size = n_features,
nlayers = args.nlayers,
res_connection = args.res_connection).to(args.device)
checkpoint = torch.load(Path('save',args.data, 'checkpoint', args.filename).with_suffix('.pth'))
model.load_state_dict(checkpoint['state_dict'])
scores, predicted_scores, precisions, recalls, f_betas = list(), list(), list(), list(), list()
targets, mean_predictions, oneStep_predictions, Nstep_predictions = list(), list(), list(), list()
try:
for channel_idx in range(n_features):
if 'means' in checkpoint.keys() and 'covs' in checkpoint.keys():
print('=> loading pre_calculated mean and covariance')
mean, cov = checkpoint['means'][channel_idx], checkpoint['covs'][channel_idx]
else:
print('=> calculating mean and covariance')
mean, cov = fit_norm_distribution_param(args, model, train_dataset, channel_idx=channel_idx)
if args.compensate == True:
print('=> training an SVR as anomaly score predictor')
train_score,_,_,hiddens,_ = anomalyScore(args,model,train_dataset,mean,cov,channel_idx=channel_idx)
score_predictor = GridSearchCV(SVR(), cv=5, param_grid={'C': [1e0, 1e1, 1e2], 'gamma': np.logspace(-1,1,3)})
score_predictor.fit(torch.cat(hiddens,dim=0).numpy(), train_score.cpu().numpy())
else:
score_predictor=None
print('calculating anomaly scores')
score, sorted_prediction, sorted_error, _, predicted_score = anomalyScore(args,model,test_dataset,mean,cov,
score_predictor=score_predictor,
channel_idx=channel_idx)
print('calculating precision, recall, and f_beta')
precision, recall, f_beta = get_precision_recall(args, score, num_samples=1000, beta=args.beta,
label=TimeseriesData.testLabel.to(args.device),
predicted_score=predicted_score)
print('data: ',args.data,' filename: ',args.filename,
' f-beta (no compensation): ', f_beta.max().item(),' beta: ',args.beta)
if args.compensate == True:
precision, recall, f_beta = get_precision_recall(args, score, num_samples=1000, beta=args.beta,
label=TimeseriesData.testLabel.to(args.device),
predicted_score=predicted_score)
print('data: ',args.data,' filename: ',args.filename,
' f-beta (compensation): ', f_beta.max().item(),' beta: ',args.beta)
target = reconstruct(test_dataset.cpu()[:, 0, channel_idx],
TimeseriesData.mean[channel_idx],
TimeseriesData.std[channel_idx]).numpy()
# 실제 값
mean_prediction = reconstruct(sorted_prediction.mean(dim=1).cpu(),
TimeseriesData.mean[channel_idx],
TimeseriesData.std[channel_idx]).numpy()
# 바로 앞 값으로 예측한 결과
oneStep_prediction = reconstruct(sorted_prediction[:, -1].cpu(),
TimeseriesData.mean[channel_idx],
TimeseriesData.std[channel_idx]).numpy()
# 10칸 앞 값으로 예측한 결과
Nstep_prediction = reconstruct(sorted_prediction[:, 0].cpu(),
TimeseriesData.mean[channel_idx],
TimeseriesData.std[channel_idx]).numpy()
sorted_errors_mean = sorted_error.abs().mean(dim=1).cpu()
sorted_errors_mean *= TimeseriesData.std[channel_idx]
sorted_errors_mean = sorted_errors_mean.numpy()
score = score.cpu()
scores.append(score), targets.append(target), predicted_scores.append(predicted_score)
mean_predictions.append(mean_prediction), oneStep_predictions.append(oneStep_prediction)
Nstep_predictions.append(Nstep_prediction)
precisions.append(precision), recalls.append(recall), f_betas.append(f_beta)
if args.save_fig:
save_dir = Path('result',args.data,args.filename).with_suffix('').joinpath('fig_detection')
save_dir.mkdir(parents=True,exist_ok=True)
plt.plot(precision.cpu().numpy(),label='precision')
plt.plot(recall.cpu().numpy(),label='recall')
plt.plot(f_beta.cpu().numpy(), label='f1')
plt.legend()
plt.xlabel('Threshold (log scale)')
plt.ylabel('Value')
plt.title('Anomaly Detection on ' + args.data + ' Dataset', fontsize=18, fontweight='bold')
plt.savefig(str(save_dir.joinpath('fig_f_beta_channel'+str(channel_idx)).with_suffix('.png')))
plt.close()
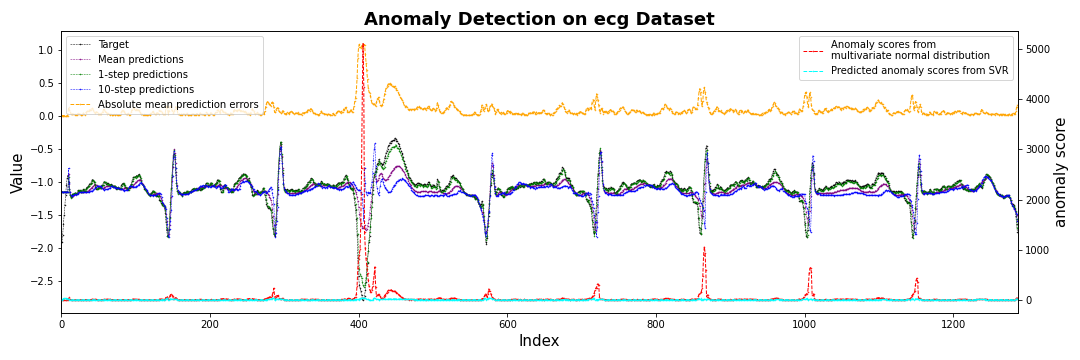
fig, ax1 = plt.subplots(figsize=(15,5))
ax1.plot(target,label='Target',
color='black', marker='.', linestyle='--', markersize=1, linewidth=0.5)
ax1.plot(mean_prediction, label='Mean predictions',
color='purple', marker='.', linestyle='--', markersize=1, linewidth=0.5)
ax1.plot(oneStep_prediction, label='1-step predictions',
color='green', marker='.', linestyle='--', markersize=1, linewidth=0.5)
ax1.plot(Nstep_prediction, label=str(args.prediction_window_size) + '-step predictions',
color='blue', marker='.', linestyle='--', markersize=1, linewidth=0.5)
ax1.plot(sorted_errors_mean,label='Absolute mean prediction errors',
color='orange', marker='.', linestyle='--', markersize=1, linewidth=1.0)
ax1.legend(loc='upper left')
ax1.set_ylabel('Value',fontsize=15)
ax1.set_xlabel('Index',fontsize=15)
ax2 = ax1.twinx()
ax2.plot(score.numpy().reshape(-1, 1), label='Anomaly scores from \nmultivariate normal distribution',
color='red', marker='.', linestyle='--', markersize=1, linewidth=1)
if args.compensate == True:
ax2.plot(predicted_score, label='Predicted anomaly scores from SVR',
color='cyan', marker='.', linestyle='--', markersize=1, linewidth=1)
#ax2.plot(score.reshape(-1,1)/(predicted_score+1),label='Anomaly scores from \nmultivariate normal distribution',
# color='hotpink', marker='.', linestyle='--', markersize=1, linewidth=1)
ax2.legend(loc='upper right')
ax2.set_ylabel('anomaly score',fontsize=15)
#plt.axvspan(2830,2900 , color='yellow', alpha=0.3)
plt.title('Anomaly Detection on ' + args.data + ' Dataset', fontsize=18, fontweight='bold')
plt.tight_layout()
plt.xlim([0,len(test_dataset)])
plt.savefig(str(save_dir.joinpath('fig_scores_channel'+str(channel_idx)).with_suffix('.png')))
#plt.show()
plt.close()
except KeyboardInterrupt:
print('-' * 89)
print('Exiting from training early')
print('=> saving the results as pickle extensions')
save_dir = Path('result', args.data, args.filename).with_suffix('')
save_dir.mkdir(parents=True, exist_ok=True)
pickle.dump(targets, open(str(save_dir.joinpath('target.pkl')),'wb'))
pickle.dump(mean_predictions, open(str(save_dir.joinpath('mean_predictions.pkl')),'wb'))
pickle.dump(oneStep_predictions, open(str(save_dir.joinpath('oneStep_predictions.pkl')),'wb'))
pickle.dump(Nstep_predictions, open(str(save_dir.joinpath('Nstep_predictions.pkl')),'wb'))
pickle.dump(scores, open(str(save_dir.joinpath('score.pkl')),'wb'))
pickle.dump(predicted_scores, open(str(save_dir.joinpath('predicted_scores.pkl')),'wb'))
pickle.dump(precisions, open(str(save_dir.joinpath('precision.pkl')),'wb'))
pickle.dump(recalls, open(str(save_dir.joinpath('recall.pkl')),'wb'))
pickle.dump(f_betas, open(str(save_dir.joinpath('f_beta.pkl')),'wb'))
print('-' * 89)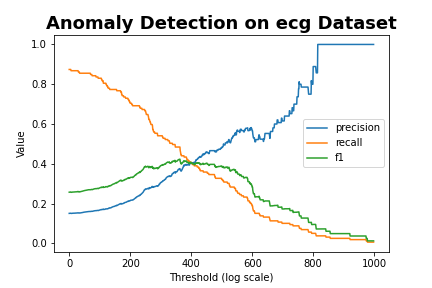
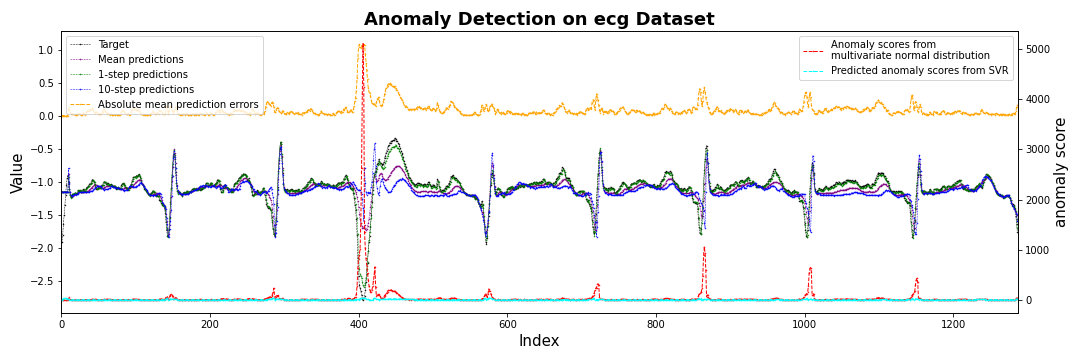
Anomaly Detection Conclusion
- f1 score가 가장 높은 350-400에서 threshold 결정
감명깊게 봤습니다. 제자가 되고 싶습니다.