Abstract
Alzheimer's disease (AD) is a devastating neurological disorder primarily affecting the elderly. An estimated 6.2 million Americans age 65 and older are suffering from Alzheimer's dementia today. Brain magnetic resonance imaging (MRI) is widely used for the clinical diagnosis of AD. In the meanwhile, medical researchers have identified 40 risk locus using single-nucleotide polymorphisms (SNPs) information from Genome-wide association study (GWAS) in the past decades. However, existing studies usually treat MRI and GWAS separately. For instance, convolutional neural networks are often trained using MRI for AD diagnosis. GWAS and SNPs are frequently used to identify genomic traits. In this study, we propose a multi-modal AD diagnosis neural network that uses both MRIs and SNPs. The proposed method demonstrates a novel way to use GWAS findings by directly including SNPs in predictive models. We test the proposed methods on the Alzheimer's Disease Neuroimaging Initiative dataset. The evaluation results show that the proposed method improves the model performance on AD diagnosis and achieves 93.5% AUC and 96.1% AP, respectively, when patients have both MRI and SNP data. We believe this work brings exciting new insights to GWAS applications and sheds light on future research directions.
[DOI] : https://doi.org/10.1109/embc46164.2021.9630174
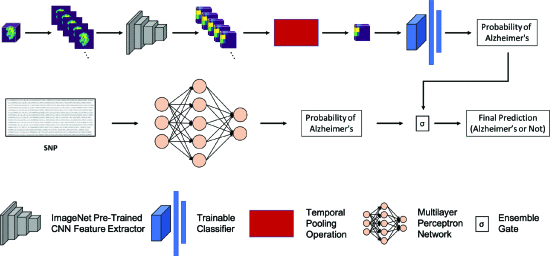
[method]
MRI- CNN(Alexnet) 이용해서 AD vs CN 분류
confidence 0.3 ~0.7 (애매할 때) 일 때 ensemble gate 열림
-> SNP 15개로 확률 측정 -> MRI probablity와 평균내서 classification
SNP
literal 증명된 15개
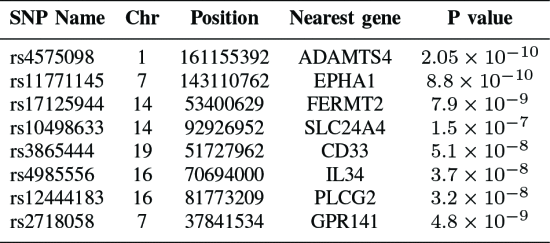
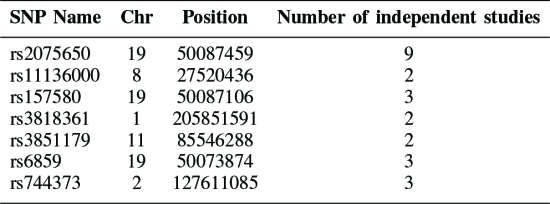
사용 SNP feature
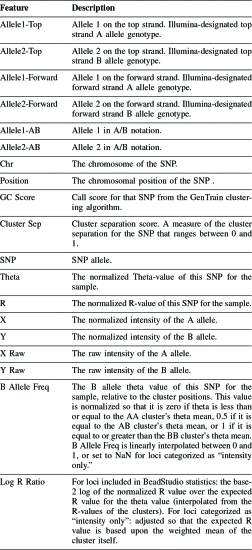
Allele1-Top, Allele2-Top, Allele1-forward, Allele2-Forward, Allele1-AB, and Allele2-AB.
MRI
-
MRI shape = H × W × K × Z
W is width of a feature map, the H is the height of a feature map, K is the number of feature maps that extracted from one slice, and Z is the number of slices in the MRI (Z ≥ 1) -
Temporal pooling operation
feature block의 slice demension 에 적용
3D -> 2D 로 전환
ouput shape = H × W × K × 1