🎛️ Data Scaling
- 데이터가 가진 크기와 편차가 다르기 때문에 한 특성을 너무 많이 반영하면 패턴을 찾아내는데 문제가 발생할 수 있음
- 이런 문제를 예방하기 위해 모든 특성의 데이터 분포나 범위를 동일하게 조정해야 함
필요한 라이브러리 호출
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.datasets import make_blobs데이터 로드
x, _ = make_blobs(
n_samples=200, centers=5,
random_state=0, cluster_std=1.5
)
train, test = train_test_split(x, random_state=5, test_size=.1)
train.shape, test.shape원본 데이터와 스케일링 된 데이터 간의 비교를 확인하기 위한 함수
def compare_scaler(train_scaled, test_scaled, scaler):
fig, ax = plt.subplots(1, 3, figsize=[20, 5])
ax[0].scatter(train[:, 0], train[:, 1], c='b', label='train data set')
ax[0].scatter(test[:, 0], test[:, 1], c='r', label='test data set')
ax[0].set_title('ORIGINAL')
ax[1].scatter(train_scaled[:, 0], train_scaled[:, 1], c='b', label='train data set')
ax[1].scatter(test_scaled[:, 0], test_scaled[:, 1], c='r', label='test data set')
ax[1].set_title(scaler)
ax[2].scatter(train[:, 0], train[:, 1], c='#B5B2FF', label='original train data set')
ax[2].scatter(test[:, 0], test[:, 1], c='#FFA7A7', label='original test data set')
ax[2].scatter(train_scaled[:, 0], train_scaled[:, 1], s=10, c='#4641D9', label='scaled train data set')
ax[2].scatter(test_scaled[:, 0], test_scaled[:, 1], s=10, c='#CC3D3D', label='scaled test data set')
plt.legend()
plt.show()◽ PowerTransformer
- 데이터의 특성별로 정규분포 형태에 가깝도록 조정
from sklearn.preprocessing import PowerTransformer
scaler = PowerTransformer()
train_scaled = scaler.fit_transform(train)
test_scaled = scaler.transform(test)
compare_scaler(train_scaled, test_scaled, 'PowerTransformer')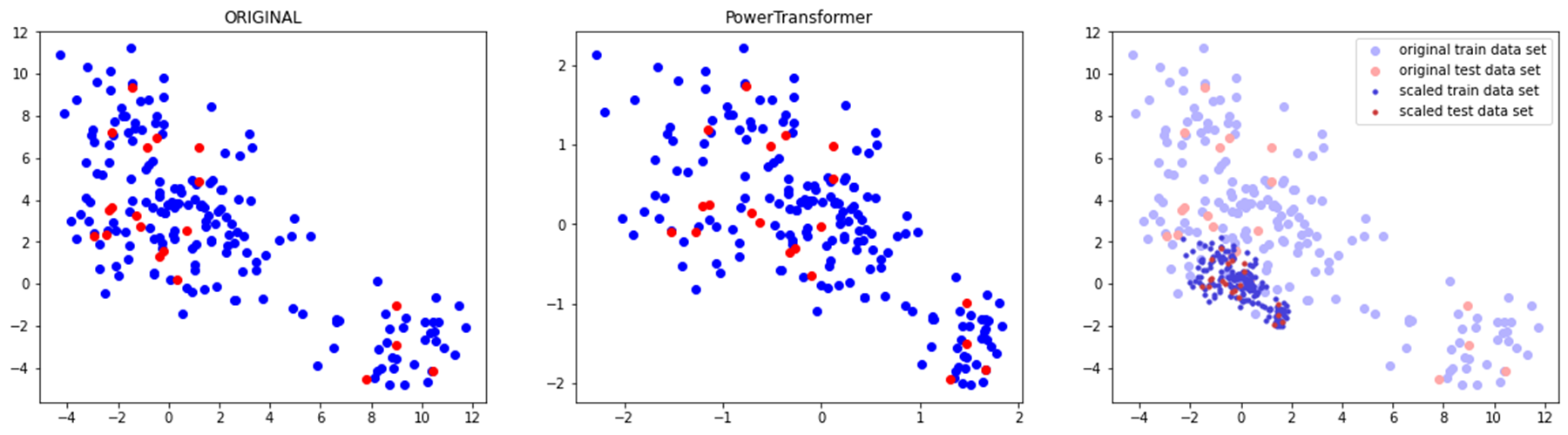
◽ QuantileTransformer
- 1000개 분위를 사용하여 데이터를 정규분포가 아닌 균등분포 시킴
(정규분포 시키려면output_distribution='normal'사용) - 이상치에 민감하지 않음
from sklearn.preprocessing import QuantileTransformer
# n_quantiles 디폴트 = 1000
scaler = QuantileTransformer(n_quantiles=train.shape[0])
# scaler = QuantileTransformer(n_quantiles=x_train.shape[0], output_distribution='normal')
train_scaled = scaler.fit_transform(train)
test_scaled = scaler.transform(test)
compare_scaler(train_scaled, test_scaled, 'QuantileTransformer')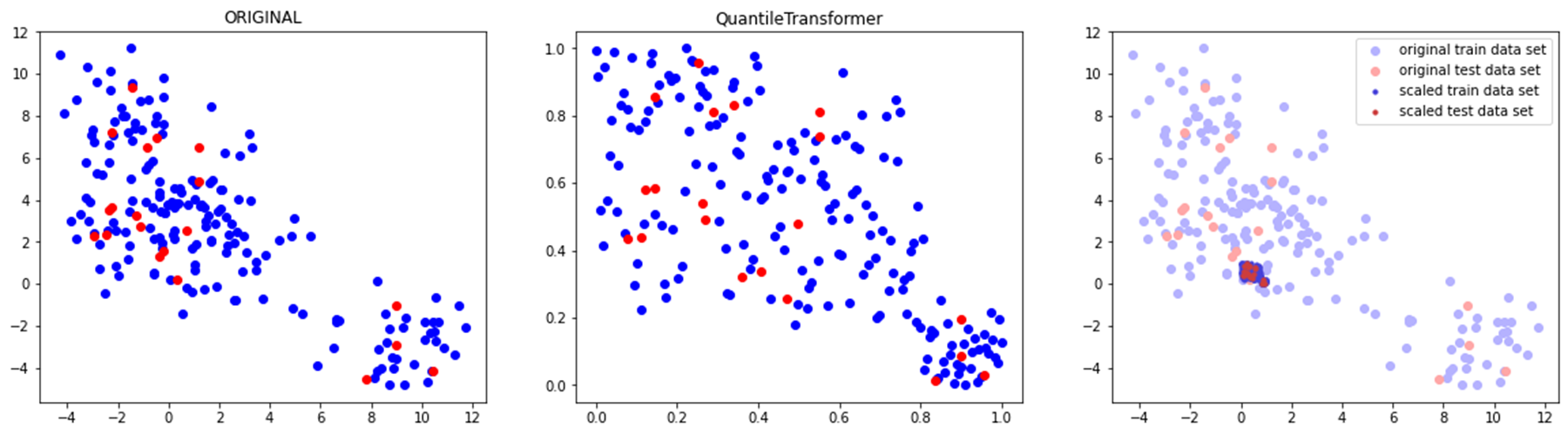
◽ MinMaxScaler
- 모든 특성들을 0과 1 사이의 값으로 조정 (최솟값 0, 최댓값 1)
- 데이터 간의 차이를 많이 줄임
- 언제 사용하나요?
- 데이터의 최솟값과 최댓값을 알 경우
- 분류보다 회귀에 유용
- 주의사항
- 이상치에 민감
- 이상치가 있을 경우 매우 좁은 범위로 압축될 수 있음
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
train_scaled = scaler.fit_transform(train)
test_scaled = scaler.transform(test)
compare_scaler(train_scaled, test_scaled, 'MinMaxScaler')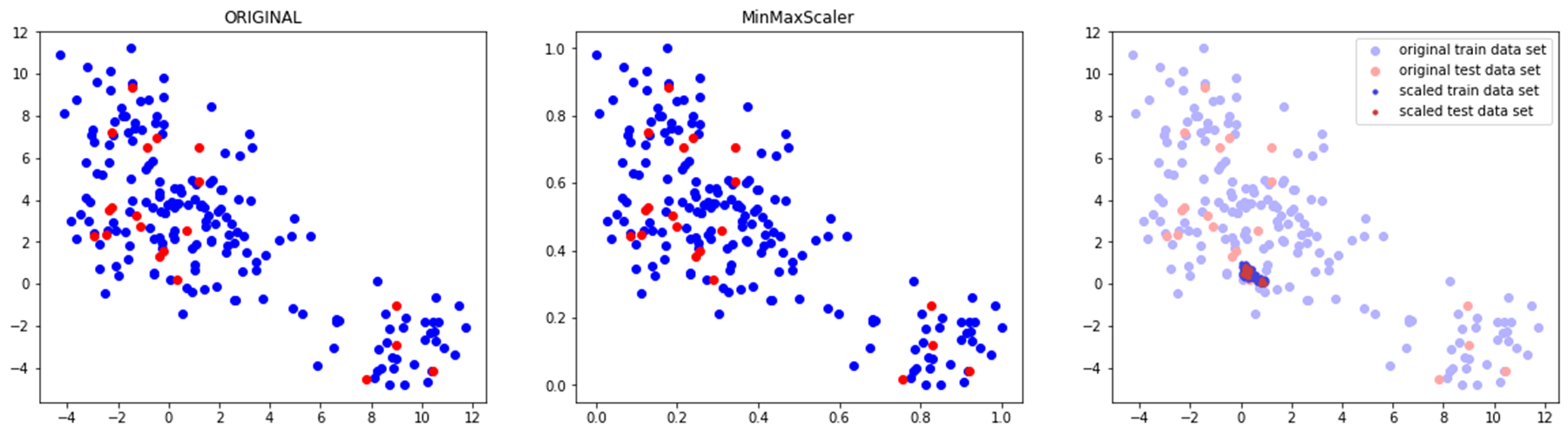
◽ MaxAbsScaler
- 특성들의 절댓값이 0과 1 사이의 값이 되도록 조정 (-1 ~ 1 사이로 조정)
- 주의사항
- 이상치가 큰 쪽에 존재할 경우 민감할 수 있음
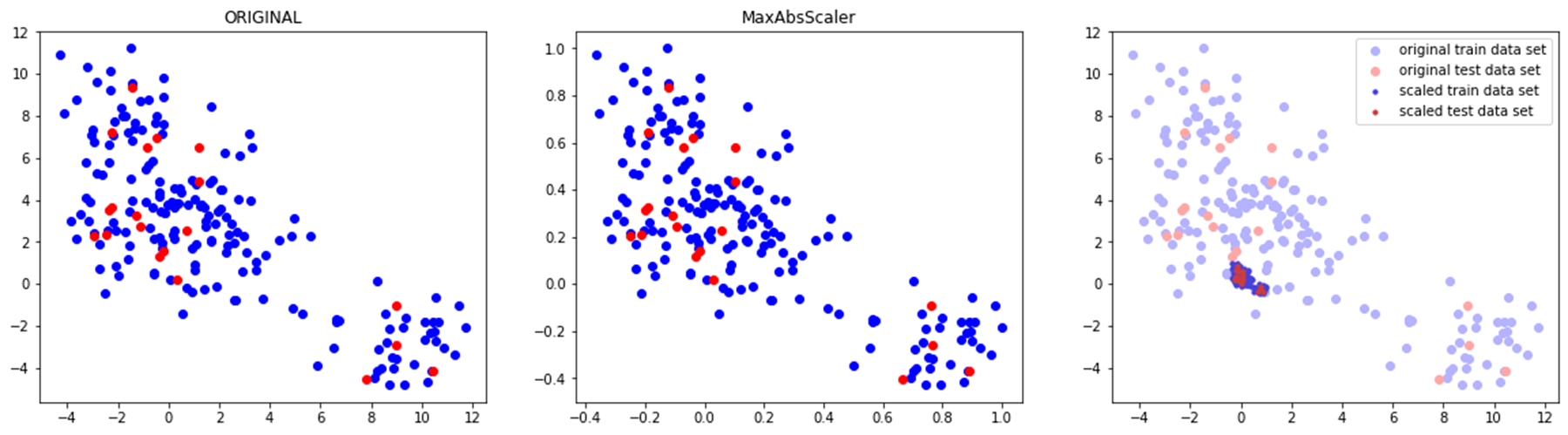
◽ StandardScaler✨
- 특성들의 평균을 0, 분산을 1로 조정 (표준정규분포에 맞춰 스케일링)
- 모든 특성들이 공통의 척도를 가짐
- 언제 사용하나요?
- 데이터의 최솟값, 최댓값을 모를 경우
- 회귀보다 분류에 유용
- 주의사항
- 이상치에 민감
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
train_scaled = scaler.fit_transform(train)
test_scaled = scaler.transform(test)
compare_scaler(train_scaled, test_scaled, 'StandardScaler')◽ RobustScaler✨
- 중앙값과 사분위값 사용
- 모든 특성들이 공통의 척도를 가짐
- 이상치에 민감하지 않음
- 언제 사용하나요?
- 이상치를 포함하는 데이터를 포함하는 경우
(이상치 영향 최소화)
- 이상치를 포함하는 데이터를 포함하는 경우
from sklearn.preprocessing import RobustScaler
scaler = RobustScaler()
train_scaled = scaler.fit_transform(train)
test_scaled = scaler.transform(test)
compare_scaler(train_scaled, test_scaled, 'RobustScaler')
📝 실제 사용 예제
데이터 로드
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
cancer_df = pd.DataFrame(
data=cancer.data, columns=cancer.feature_names
)
cancer_df['target'] = cancer.target
cancer_df.head()
x_train, x_test, y_train, y_test = train_test_split(
cancer.data, cancer.target,
test_size=0.2, random_state=3
)스케일러 사용 X
clf = DecisionTreeClassifier()
clf.fit(x_train, y_train)
no_scaler_score = clf.score(x_test, y_test)
print('No Scaler 모델 정확도 :', no_scaler_score)No Scaler 모델 정확도 : 0.8859649122807017스케일러 사용 O
scaler = MinMaxScaler()
train_scaled = scaler.fit_transform(x_train)
test_scaled = scaler.transform(x_test)
clf = DecisionTreeClassifier()
clf.fit(train_scaled, y_train)
scaler_score = clf.score(test_scaled, y_test)
print('MinMaxScaler 모델 정확도 :', scaler_score)MinMaxScaler 모델 정확도 : 0.9122807017543859scaler.fit_transform()fit(): 데이터 학습 함수transform(): 학습한 것을 적용하여 변환하는 함수fit()+transform()- 학습셋에 사용, 테스트셋에는 절대 사용하면 안됨
- 테스트셋에 사용할 경우 기존에 학습셋의 기준을 무시하고 새로운 데이터를 학습하는 것

