[Paper Review] SPECTER: Document-level Representation Learning using Citation-informed Transformers
Paper Review
목록 보기
1/1

SPECTER: Document-level Representation Learning using Citation-informed Transformers
URL: https://arxiv.org/pdf/2004.07180.pdf
Year: 2020
1. Abstract
- 최근 Transformer 기반한 많은 LM 모델들은 learn powerful textual representation
- 하지만 대다수의 LM 모델들은 Token or Sentence-level 에서 학습이 이루어지지만, Document-level 에서의 학습은 이루어지지 않음
- 따라서, 이 논문에서는 Transformer 기반 Scientific Document Embedding 을 위한 SPECTER* (Scientific Paper Embeddings using Citation-informed TransforERs)모델 제시 + 벤치마크 데이터 공개
2. Introduction
Background
- 최근 많은 양의 Scientific Publication → 사용자가 원하는 논문의 검색 혹은 추천에 NLP가 필요 → Hence, Science domain & Document embedding
- SPECTER 이전에도 Document Embedding의 연구가 있었지만 (Tu et al. 2017, Chen et al. 2019), BERT와 같은 SOTA pretrained LM은 적용하지 않음
Document Processing
- 논문의 Title 과 Abstract 는 많은 정보를 내포하지만 ,이를 off-the-shelf 식으로 pretrained LM에 넣어주면 큰 의미가 없다 (see later)
- 가령, Science Domain에서 SOTA를 달성한 SciBERT (BERT_base + 1.18M Paper)
- 기존 BERT 모델에 118만개의 페이퍼 (from Semantic Scholar, 18% CS, 82% BioMedic) 데이터를 Fine-tuned
- SciBERT Result
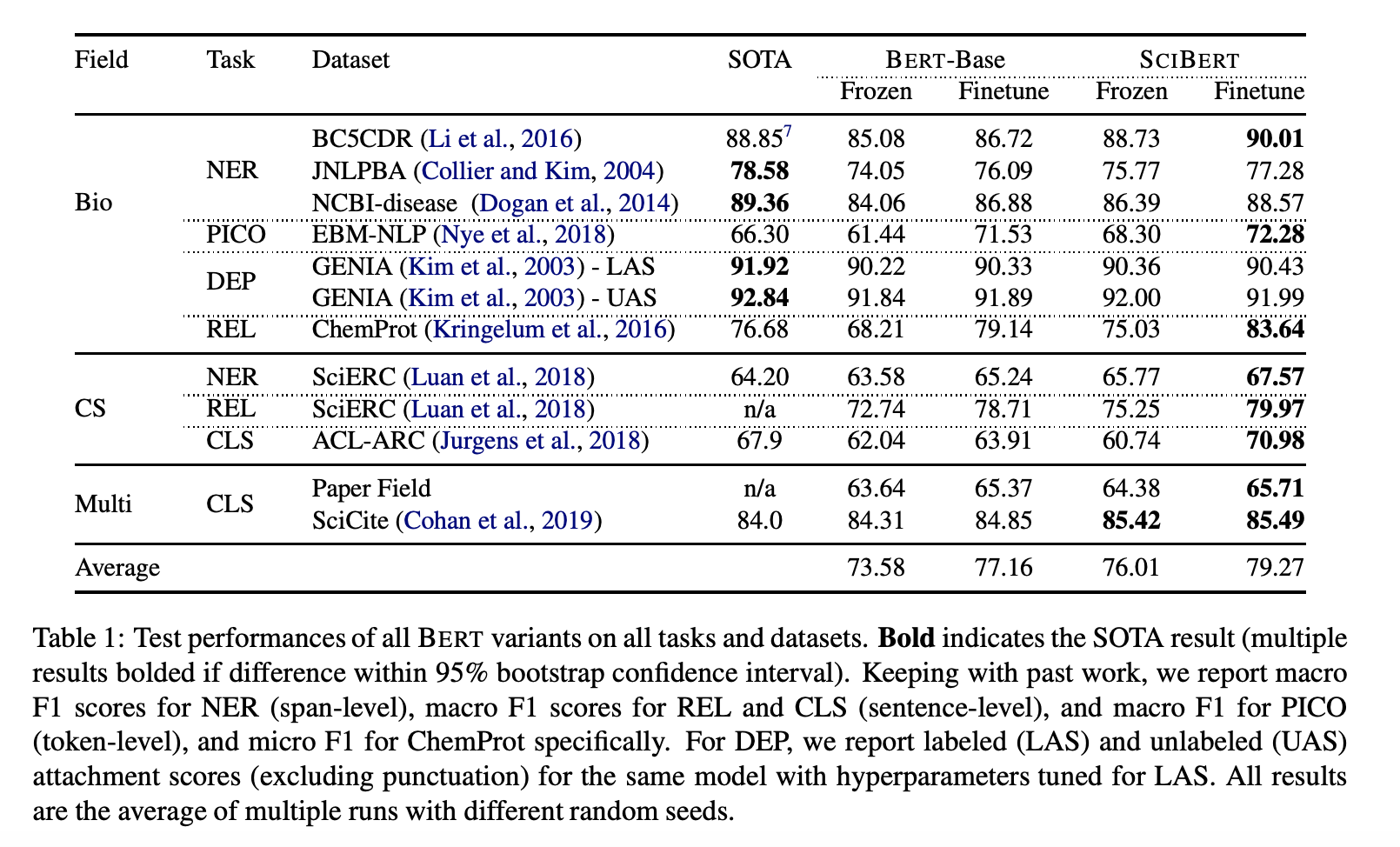
- SPECTER는 논문간의 Citation을 naturally occurring signal → Which documents are most related and apply triplet-loss for objective function
- 이를 통해 Document-level의 여러 Task에서 (topic classification, citation prediction, recommendation) 에서 SOTA
3. Model
Overview
Triplet
- Each training instance is a triplet of papers : a Query paper , a Positive paper and a Negative paper
- Positive paper : paper that is cited by Q
- Negative paper : paper that is NOT cited by Q (but could be cited by )
- Random Negative
- Choose a random paper from the corpus
- Hard Negative
- A paper which is NOT cited by Query paper but IS cited by Positive paper
- Random Negative
- Loss Function
- The L2 norm distance is used :
Encoder & Data
- Use SciBERT for foundation and fine-tune it with Semantic Scholar data
- 146K query papers for train, 32K papers for validation
- For each train Query paper → 5 distinct triplets
## Emperically found that 2 hard negatives and 3 random negatives are 'helpful' | Query | Positive | Negative | |-------|------------|-----------| | Q1 | Citation 1 | Hard_Neg1 | | Q1 | Citation 2 | Hard_Neg2 | | Q1 | Citation 3 | Rand_Neg1 | | Q1 | Citation 4 | Rand_Neg2 | | Q1 | Citation 5 | Rand_Neg3 |
- Encoder Input = SciBERT( [CLS] Title + [SEP] + Abstract)
- Use [CLS] token of the last layer for final document embedding
- During inference, no citations are required
SciDocs
Introduce a new comprehensive evaluation framework to measure the effectiveness of scientific paper embeddings (SCIDOCS)
- Document Classification (F1-score)
- Medical Subject Headings (MeSH) Classification → 11 classes
- Paper Topic Classification → 19 classes
- Citation Prediction (Ranking, MAP & nDCG score)
- Direct Citation
- Predict which papers are cited by a given query paper from a given set of candidate papers (5 cited + 25 random)
- Co-Ciation
- Predict which papers are highly co-cited with a given query paper from a given set of candidate papers
- Direct Citation
- User-Activitiy (Ranking, MAP & nDCG score)
- Co-Views
- Predict which papers are viewed within the same browser
- Co-Reads (Ranking, P@1 score)
- Predict which papers are read while a user is reading a query paper
- Co-Views
- Recommendation
- Based on user's clickthrough data, rank a given set of candidate papers
For details of MAP and nDCG score, please see this link
4. Result
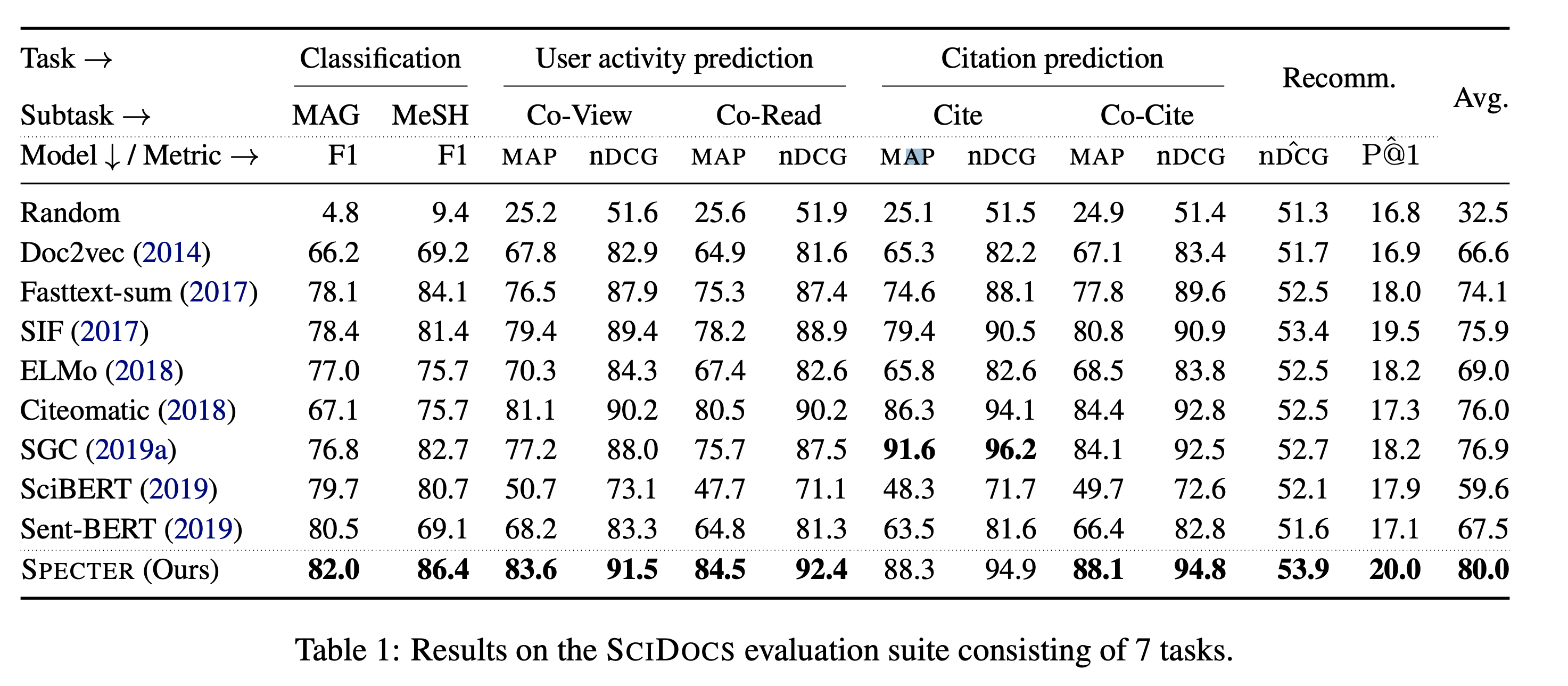
- All experiments are performed in feature-based fashion, as opposed to fine-tuned
- For Classification, an extra SVM layer is added
- For User-activity & Citation prediction, direct L2 distance is calculated
- For Recommendation, an extra Feed-Forward network is added (in case where extra features such as authors, dates and venues are added)
- Overall, significant improvements on almost tasks
- Except for SGC (Simple Graph Convolution) → It has an access to Citation Graph during test time
- From the comparison with SciBERT → Citation-based Triplet Loss is critical !
Ablation Study
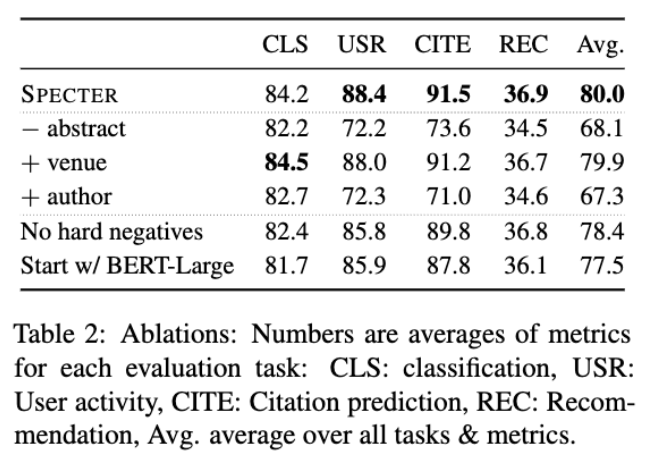
- Removing Abstract degrades performance
- Adding meta-data such as Author and Venue also degrades performance
- Author → likely to be OOV for SciBERT Wordpieces tokenizer
- Venue → Gives huge information on topic (eg. ICML, EMNLP)
- Hard Negatives are helpful
- BERT-LARGE performs worse than SciBERT for initial encoder
Feature-Based vs Fine-Tuned
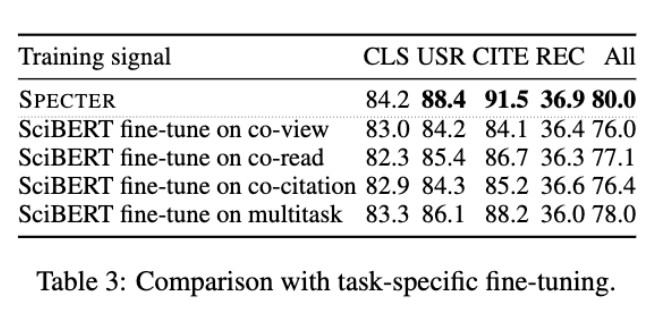
- SPECTER works significantly better than Fine-Tuned SciBERT
5. Conclusion
- Document representation learning with pre-trained LM (SciBERT)
- Apply inter-document relatedness (i.e. citation) for document embedding
- Embedding based on only the title and abstract
- Could have improved accuracy with FULL text
- But often full text is not available + Memory issue
- Release of new benchmark dataset - SciDoc
