

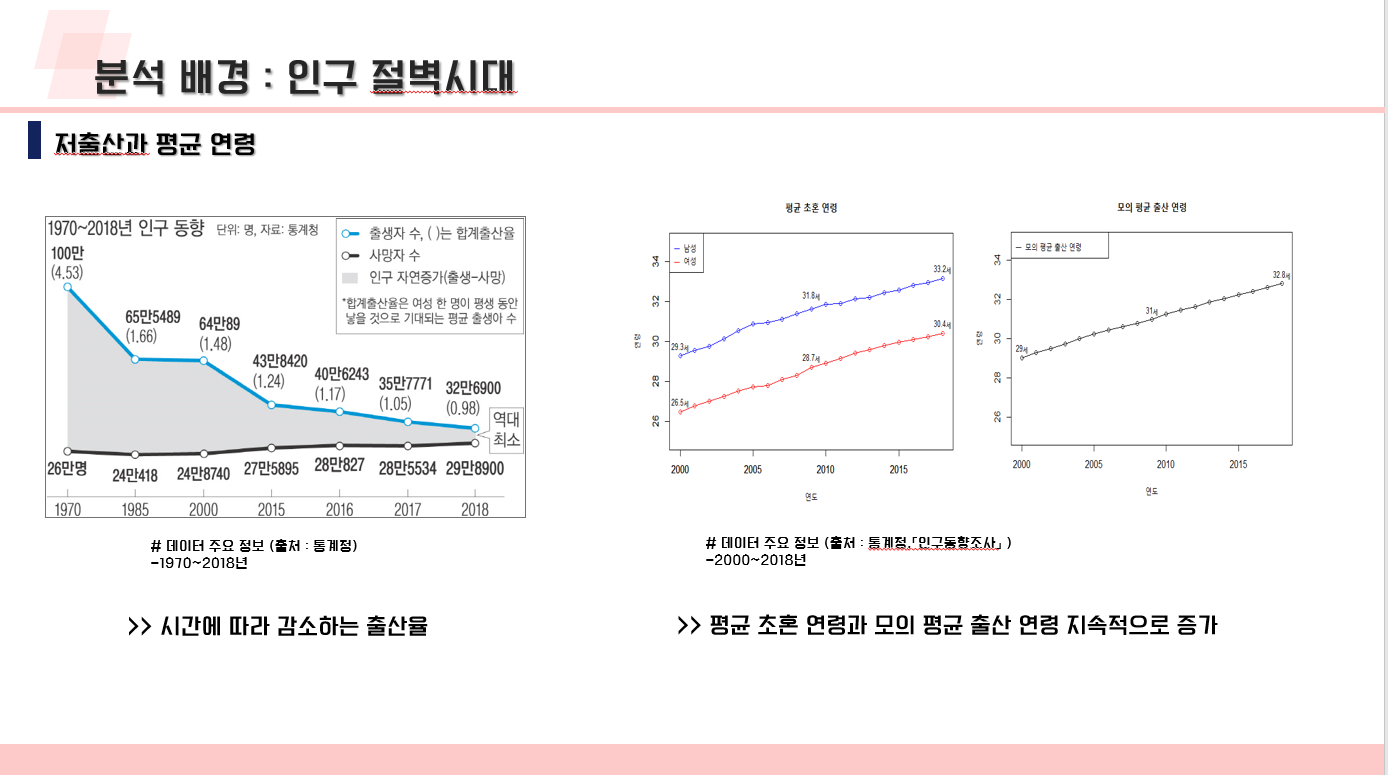
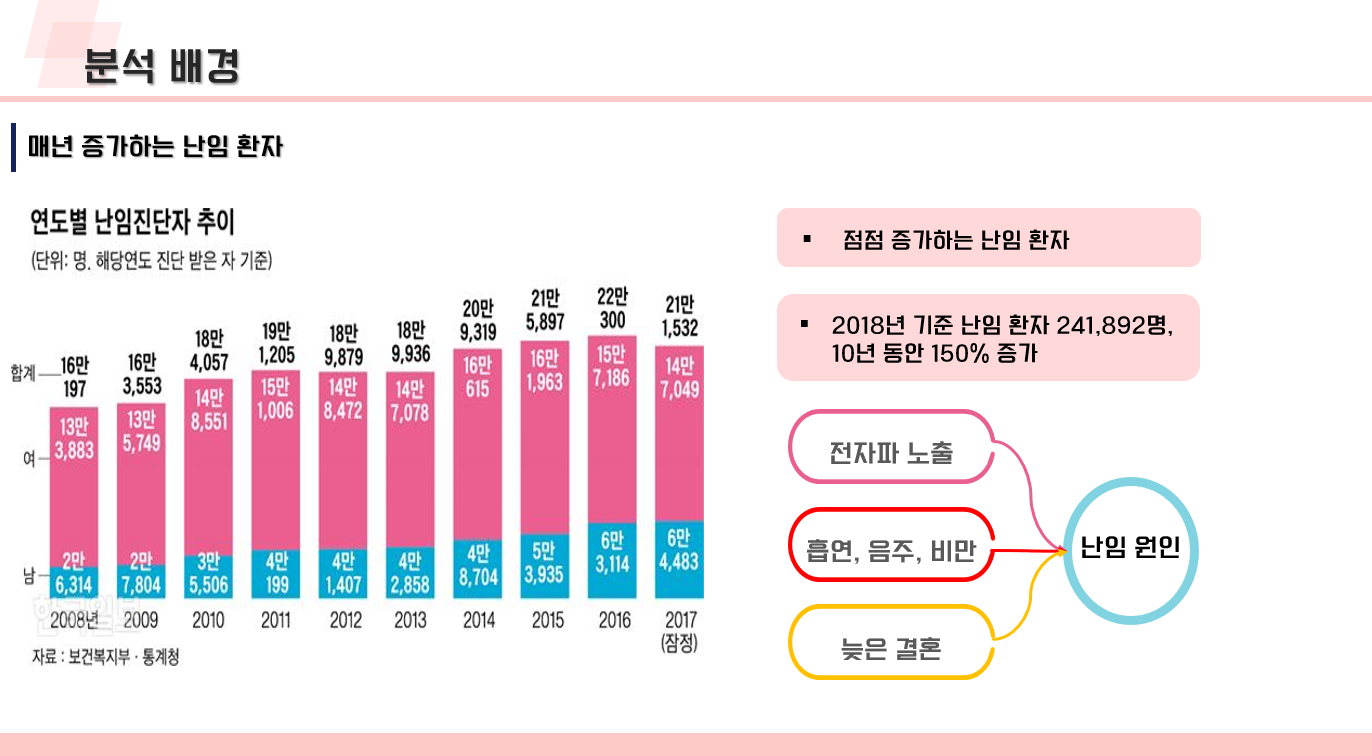
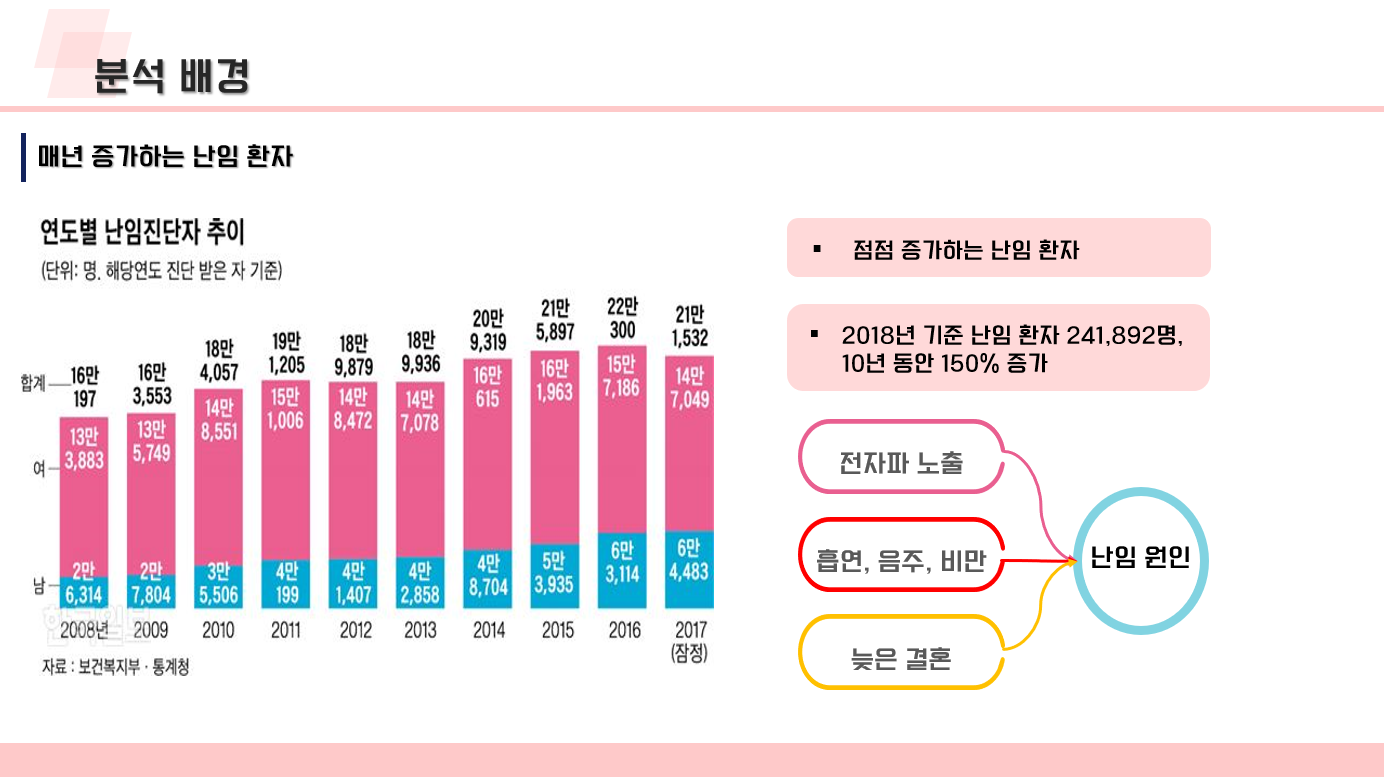
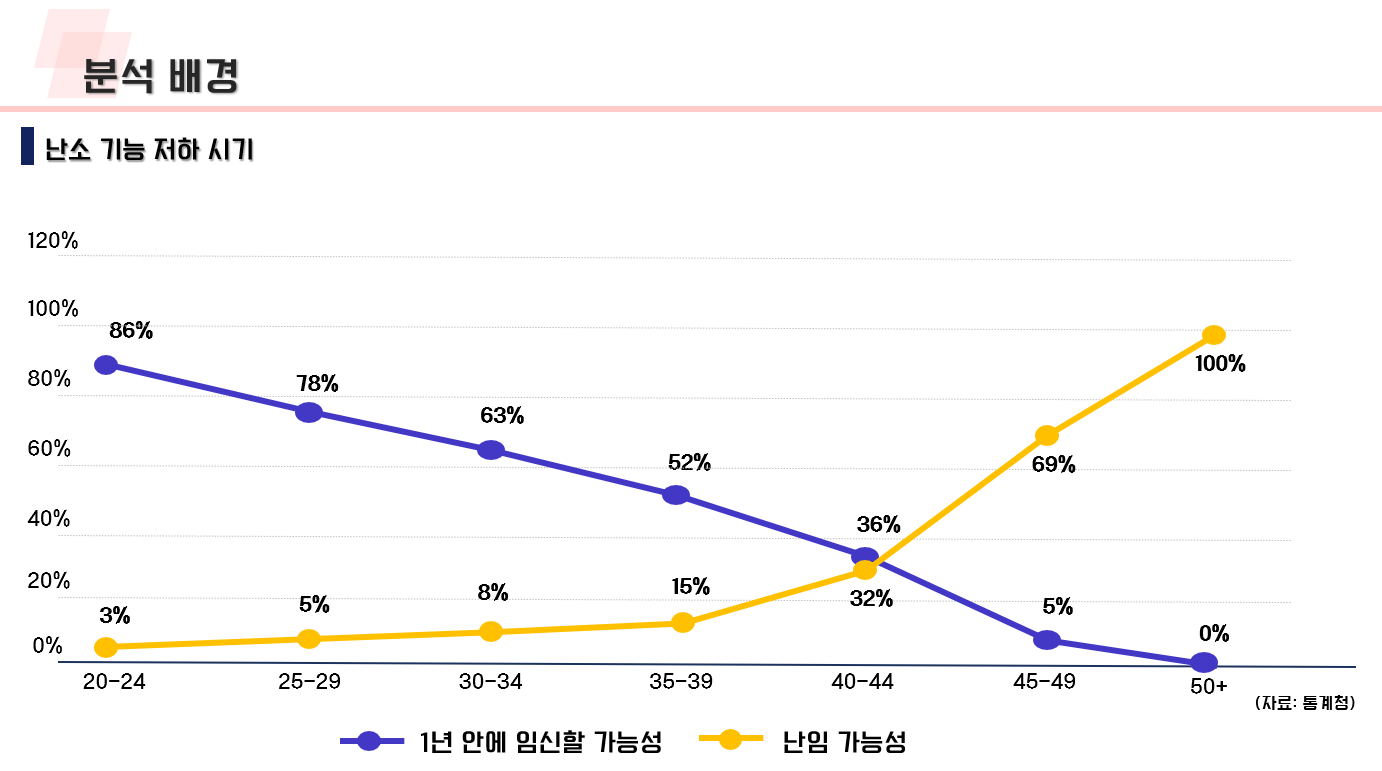
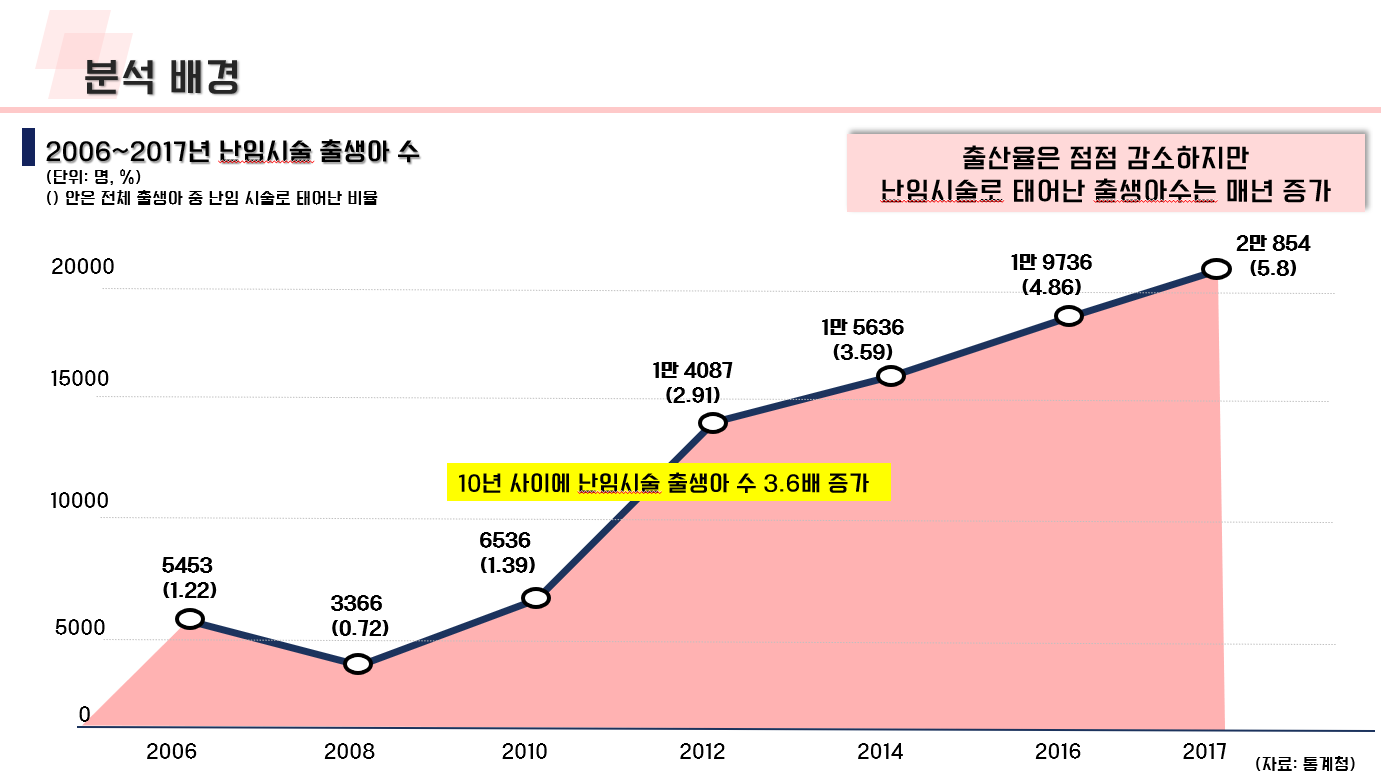
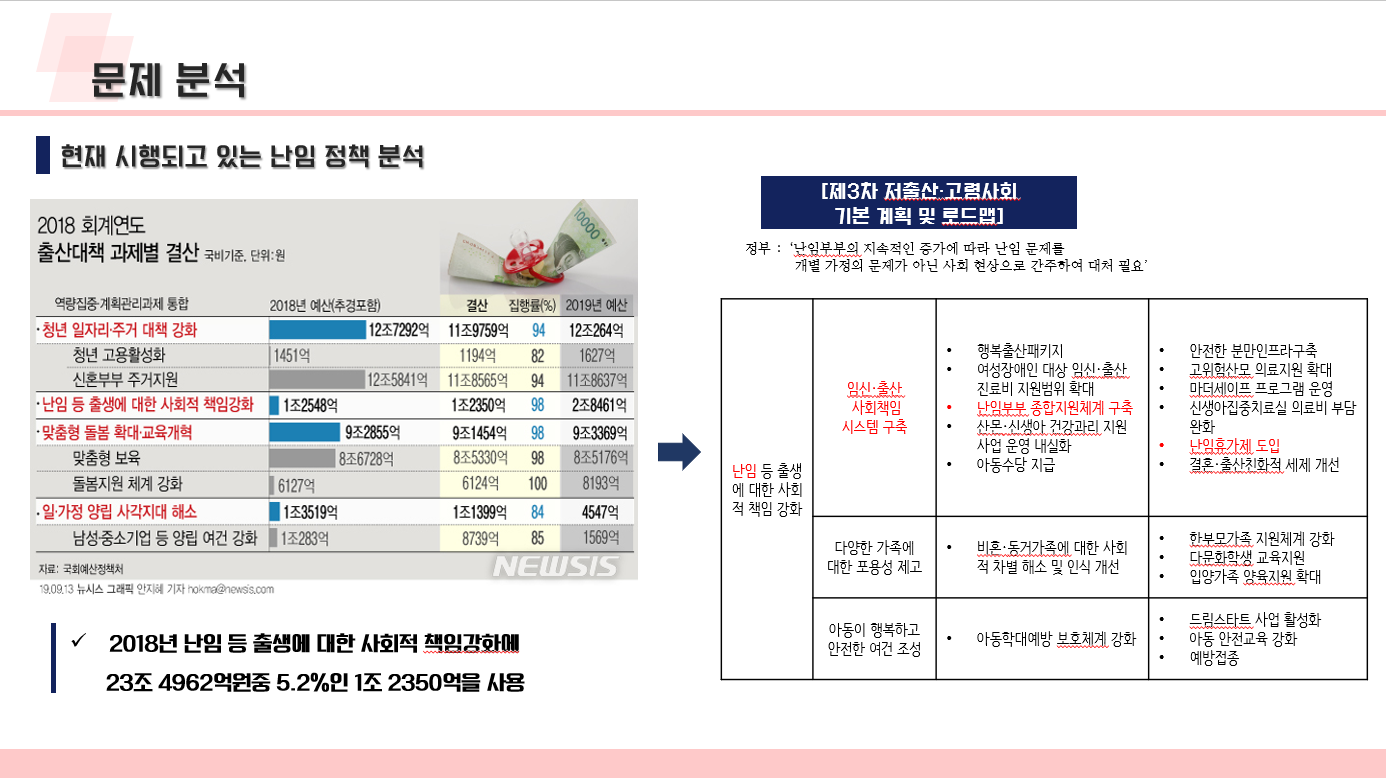


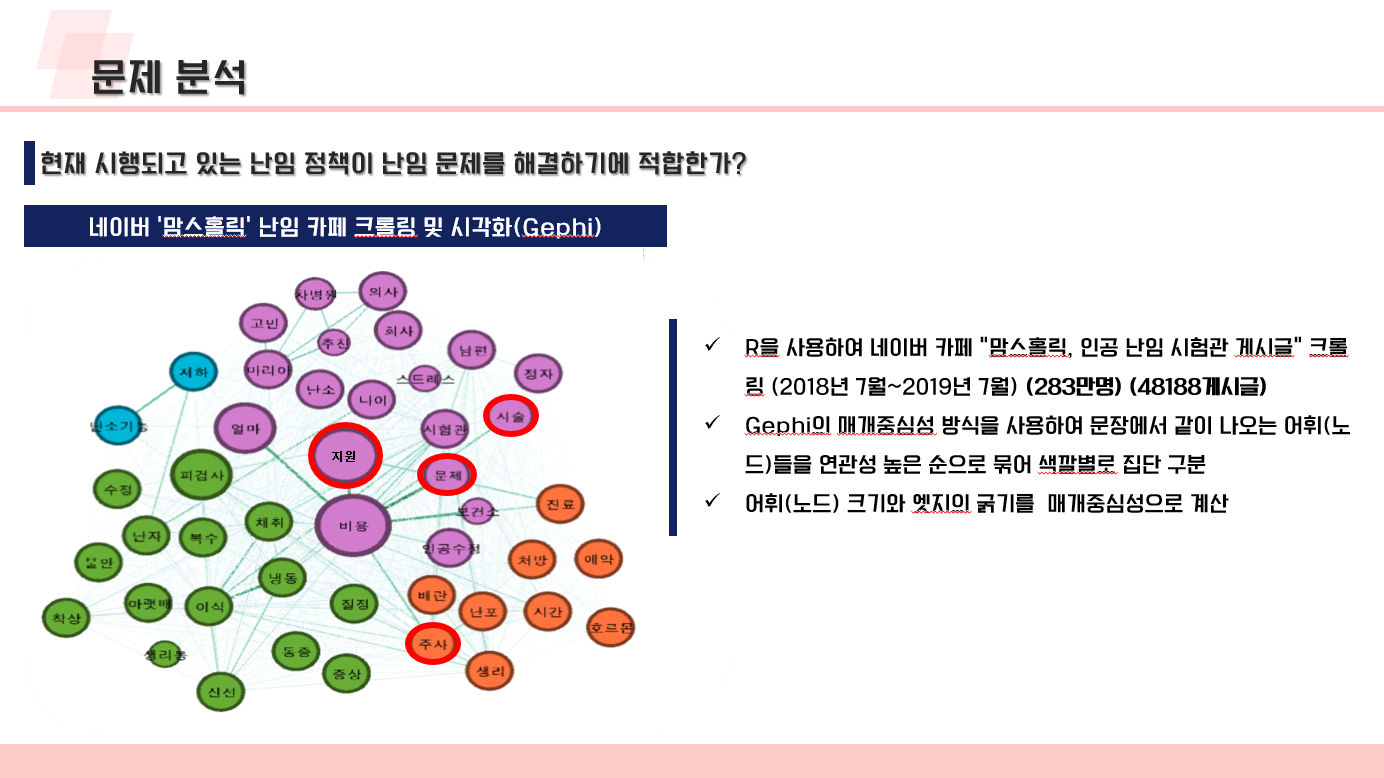
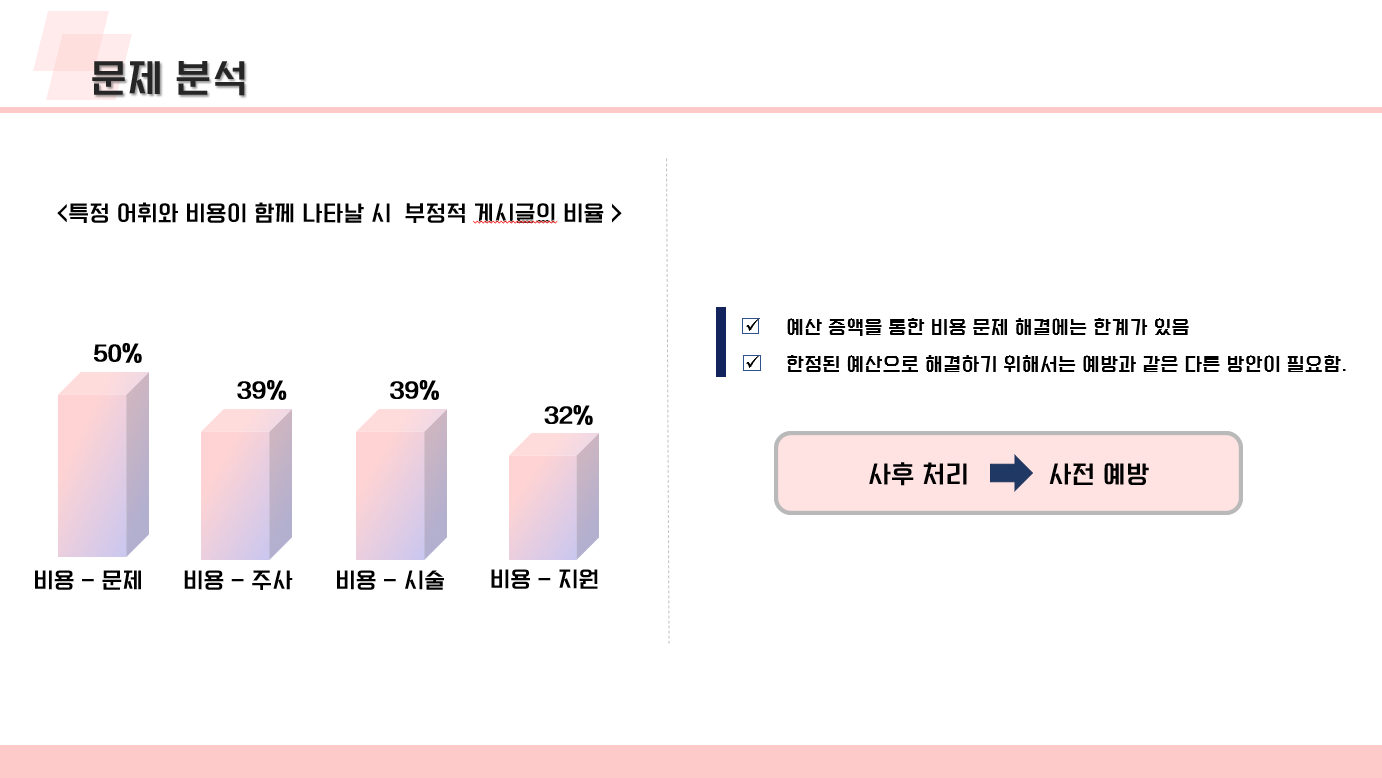


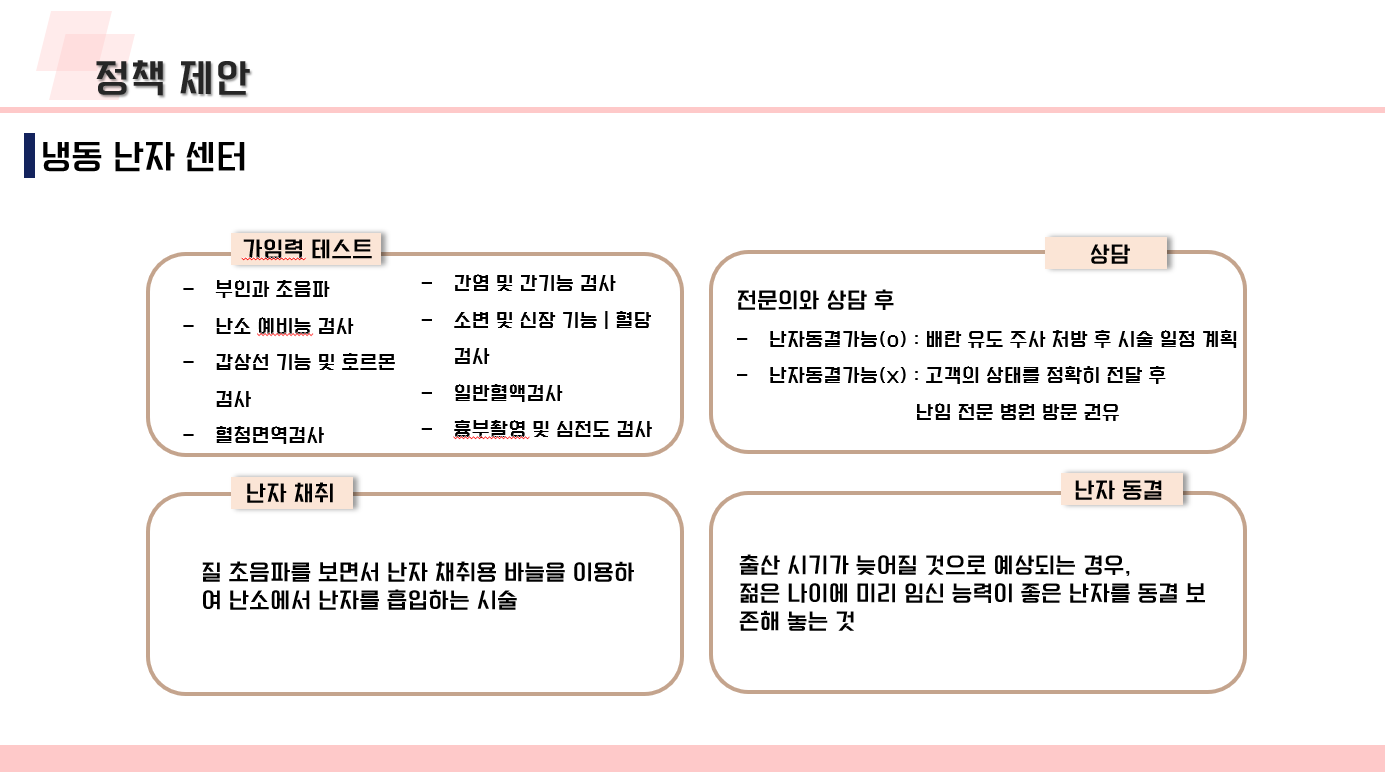
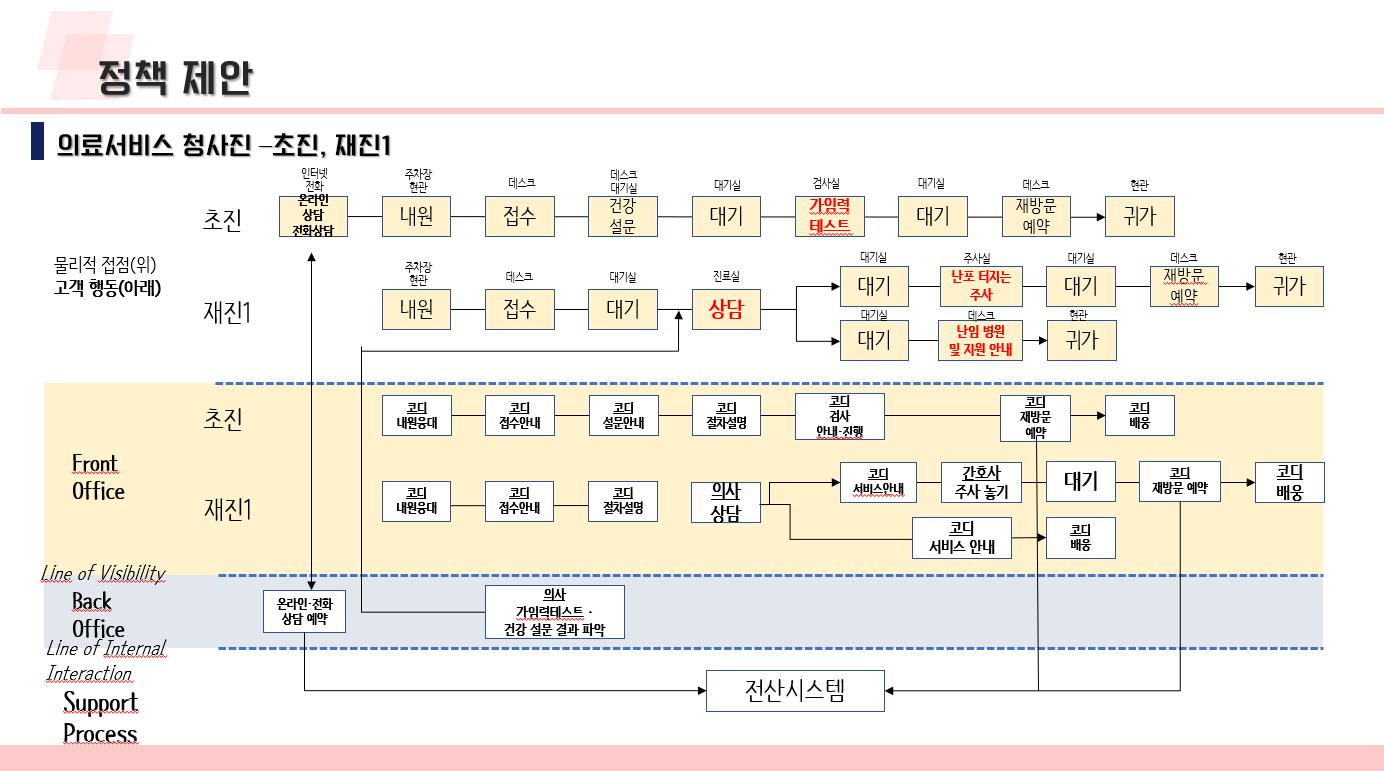
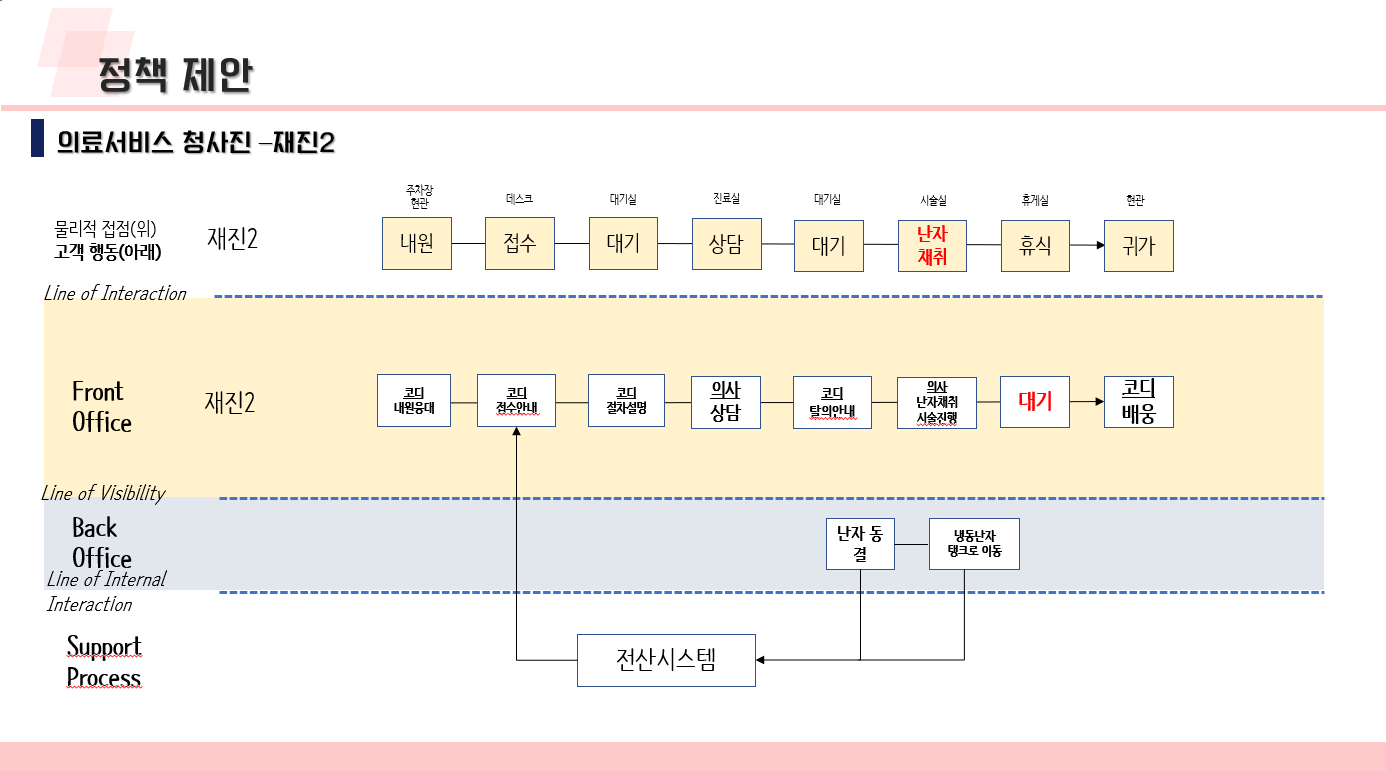
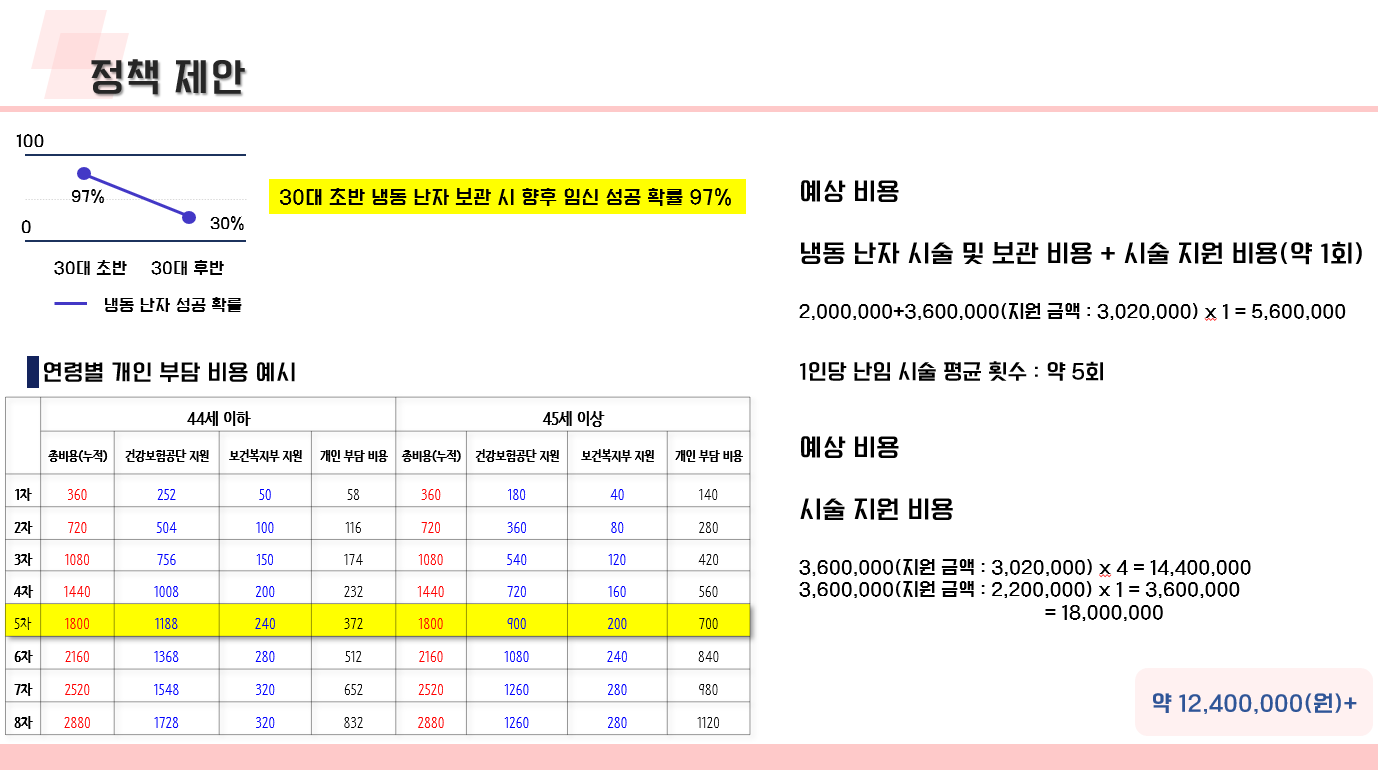
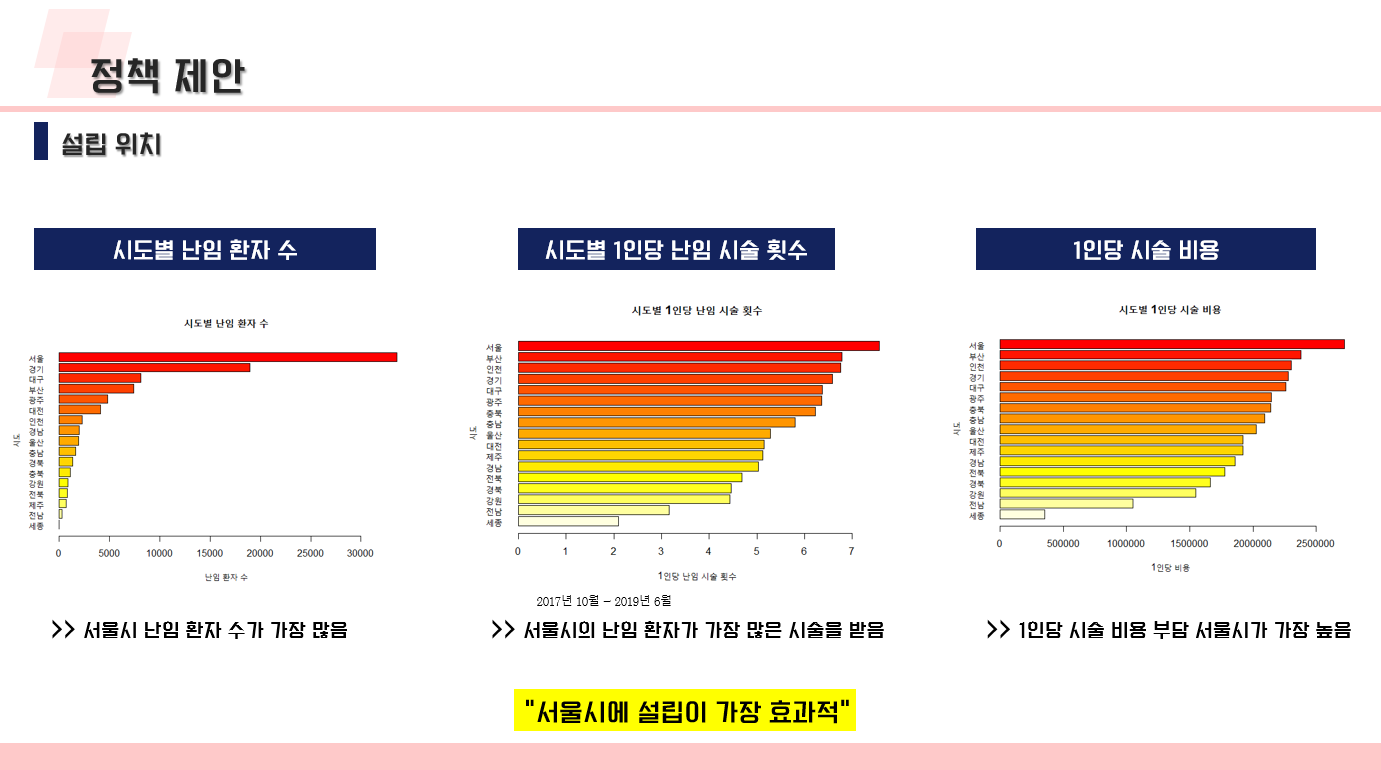

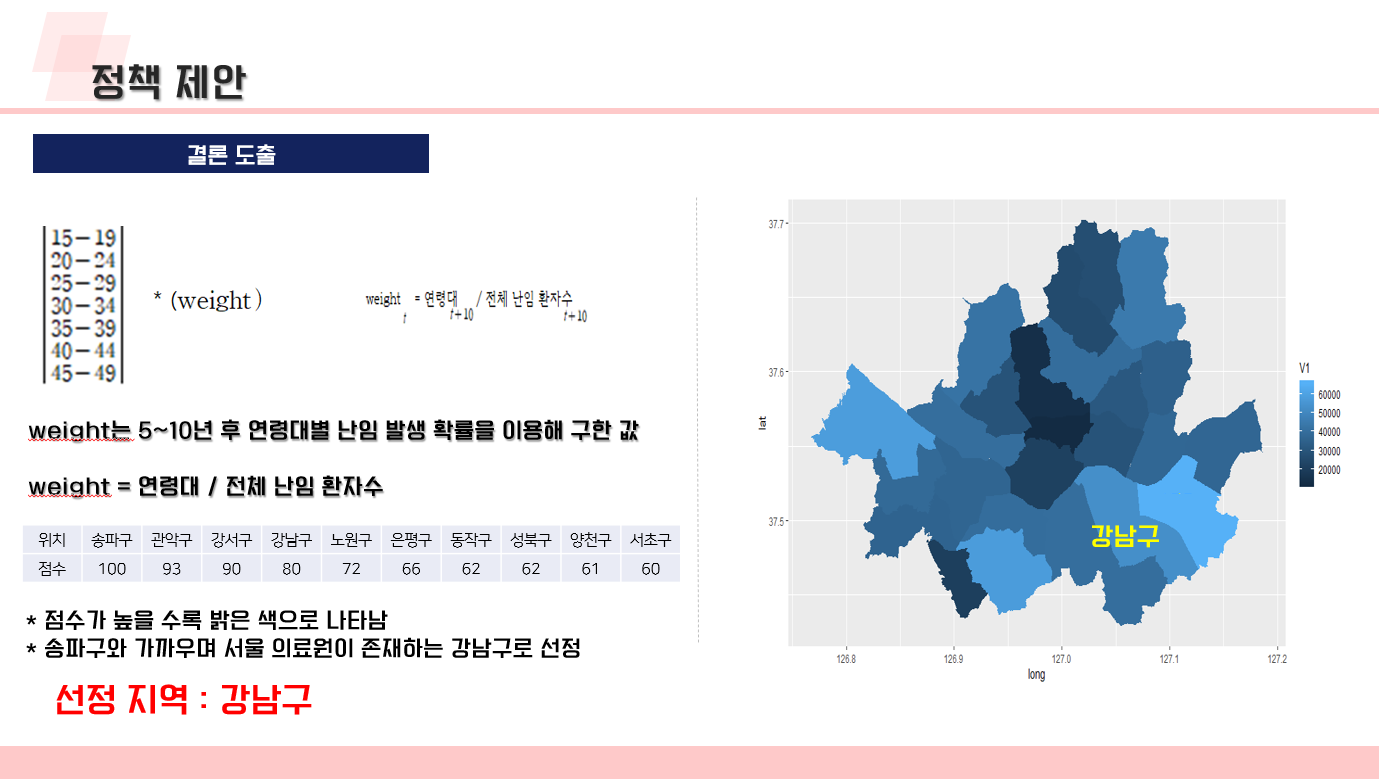
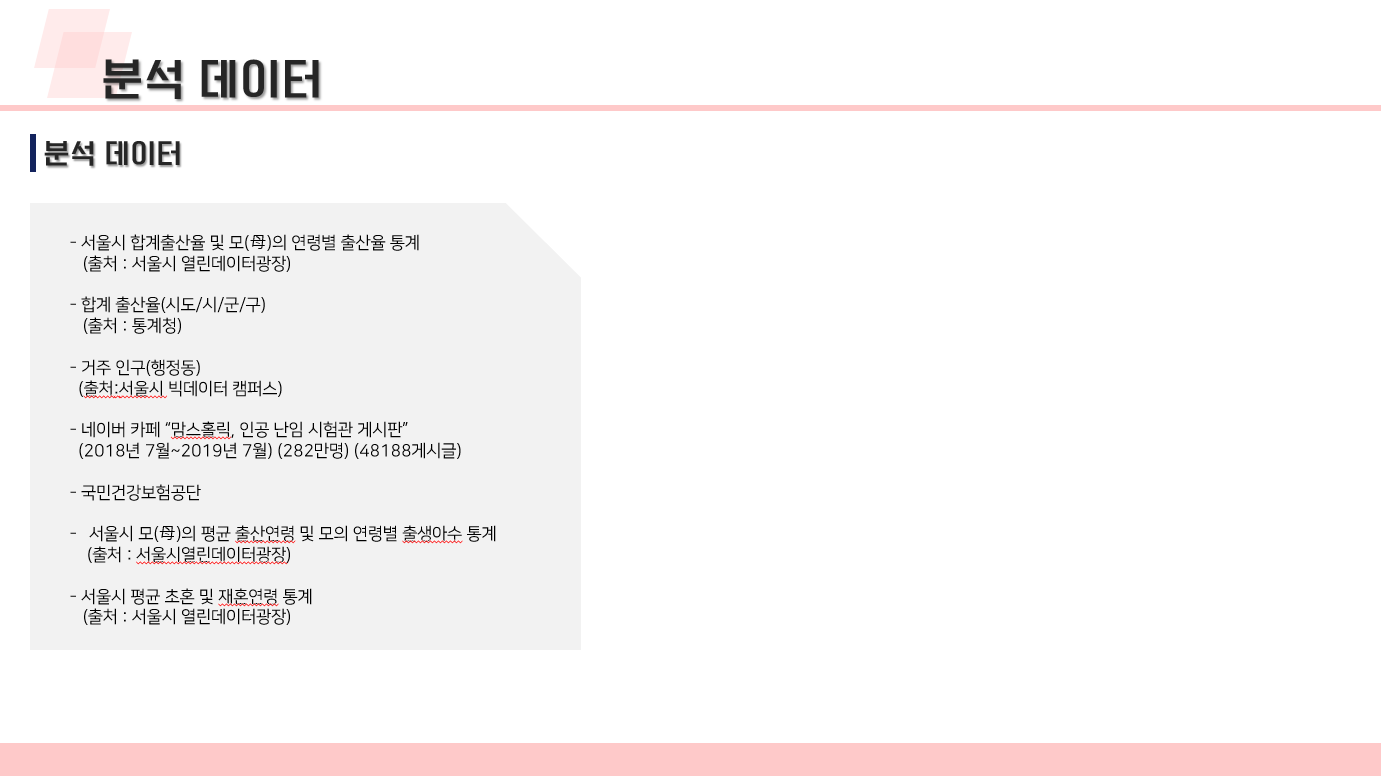


난임 관련 데이터 수집 코드
#################################################################################
# 패키지 설치 및 실행
# install.packages("RSelenium")
# install.packages("rvest")
# install.packages("httr")
# install.packages("stringr")
# install.packages("dplyr")
# install.packages("KoNLP")
# install.packages("RColorBrewer")
# install.packages("wordcloud")
library(KoNLP)
library(RSelenium)
library(rvest)
library(httr)
library(stringr)
library(dplyr)
library(RColorBrewer)
library(wordcloud)
#################################################################################
# 스크래이핑
# 포트번호 할당
portn <- as.integer(runif(1, 1,5000))
# 드라이버 설정
rD <- rsDriver(port=portn, browser="chrome", chromever = "75.0.3770.8")
# 클라이언트 드라이버 이름 설정
remDr <- rD[["client"]]
# 네이버로그인 화면으로 이동
remDr$navigate("https://section.cafe.naver.com/")
# 버튼 위치 찾기
btn <- remDr$findElement(using="css selector", value="#gnb_login_button > span.gnb_txt")
# 로그인 버튼 클릭
btn$clickElement()
# 아이디와 비밀번호 찾기
id <- remDr$findElement(using="id", value="id")
pw <- remDr$findElement(using="id", value="pw")
# 입력란에 아이디와 비밀번호 입력하기
id$setElementAttribute("value", "chapr1")
pw$setElementAttribute("value", "ckzpdjtm")
# ID 방식으로 버튼 위치 찾기
btn <- remDr$findElement(using="class", value="btn_global")
# 로그인 버튼 클릭
btn$clickElement()
# 불임은 없다. 맘스홀릭 페이지로 이동
remDr$navigate("https://cafe.naver.com/ArticleList.nhn?search.clubid=10094499&search.menuid=418&search.boardtype=L")
# iframe 으로 들어가기
iframe <- remDr$findElement(using = "xpath", value = '//*[@id="cafe_main"]')
remDr$switchToFrame(iframe)
# 페이지 전체 소스 가져오기
frontPage <- remDr$getPageSource()
# URL 수집
html <- read_html(frontPage[[1]])
cafeURL <- html %>% html_nodes("a") %>% html_attr("href")
cafeURL <- cafeURL[grepl("articleid=", cafeURL) == TRUE & grepl("referrerAllArticles=", cafeURL) == TRUE &
grepl("commentFocus=true", cafeURL) == FALSE]
cafeDate <- html %>% html_nodes(".td_date") %>% html_text()
# 공지날짜만 제거
cafeDate <- cafeDate[-(1:21)]
# 최대 페이지 지정
tnum <- 1000
for (i in 1:tnum) { # i <- 288
j <- ifelse(i > 11, i - 9 - 10*((i - 2)%/%10 - 1), i)
print(c(i,j))
# 버튼 element 찾기
webElem <- remDr$findElements(using = 'xpath', value = paste0('//*[@id="main-area"]/div[7]/a[',j,']'))
if(length(webElem) == 0){
}else{
webElem[[1]]$clickElement()
# 잠시 동작 멈춤
Sys.sleep(1)
# 페이지 전체 소스 가져오기
frontPage <- remDr$getPageSource()
html <- read_html(frontPage[[1]])
cafeURLTemp <- html %>% html_nodes("a") %>% html_attr("href")
cafeURLTemp <- cafeURLTemp[grepl("articleid=", cafeURLTemp) == TRUE & grepl("referrerAllArticles=", cafeURLTemp) == TRUE &
grepl("commentFocus=true", cafeURLTemp) == FALSE]
cafeDateTemp <- html %>% html_nodes(".td_date") %>% html_text()
# 데이터 병합
cafeURL <- append(cafeURL, cafeURLTemp)
cafeDate <- append(cafeDate, cafeDateTemp)
}
}
# cafeURL
# cafeDate
cafeDate1 <- cafeDate
cafeDate1[nchar(cafeDate1) == 5] <- "2019.07.31."
cafeDate2 <- cafeDate1
cafeDate2 <- substr(cafeDate2,1,8) == "2019.07."
table(cafeDate2)
cafeURL2 <- cafeURL
cafeURL2 <- cafeURL2[cafeDate2]
# 컨텐츠 수집
cafe_gen <- list()
cafe_content <- list()
for ( i in 1:length(cafeURL2)){ # i <- 2
print(i)
n_url <- paste0("https://cafe.naver.com", cafeURL2[[i]])
remDr$navigate(n_url)
rr <- try(remDr$getPageSource(), silent=TRUE)
if(grepl("try-error", rr)){
remDr$dismissAlert()
date1 <- ""
x <- ""
cafe <- cbind(date= date1, url = n_url)
cafe_gen[[length(cafe_gen)+1]] <- cafe
cafe_content[[length(cafe_content)+1]] <- x
}else{
iframe <- remDr$findElement(using = "xpath", value = '//*[@id="cafe_main"]')
remDr$switchToFrame(iframe)
frontPage <- remDr$getPageSource()
html <- read_html(frontPage[[1]])
date <- html_nodes(html, xpath='//*[@class="m-tcol-c date"]')
date1 <- html_text(date)
print(date1)
html <- html_nodes(html, xpath='//*[@id="tbody"]/p')
temp <- html_text(html)
x <- gsub("[[:punct:]]", "", temp)
x <- gsub("ㅅㅇㅇㅊ", "서울역차병원", x)
x <- gsub("ㄱㄴㅊ", "강남차병원", x)
x <- gsub("ㅂㄷㅊ", "분당차병원", x)
x <- gsub("ㅅㅇㅇ차", "서울역차병원", x)
x <- gsub("ㄱㄴ차", "강남차병원", x)
x <- gsub("ㅂㄷ차", "분당차병원", x)
x <- gsub("서울역ㅊ", "서울역차병원", x)
x <- gsub("분당ㅊ", "분당차병원", x)
x <- gsub("서울역ㅊ", "서울역차병원", x)
x <- gsub("분당ㅊ", "분당차병원", x)
x <- gsub("강남ㅊ", "강남차병원", x)
x <- gsub("서울역차", "서울역차병원", x)
x <- gsub("분당차", "분당차병원", x)
x <- gsub("강남차", "강남차병원", x)
x <- gsub("ㅁㄹㅇ", "마리아", x)
x <- gsub("ㅅㅅㅁㄹㅇ", "신설마리아", x)
x <- gsub("ㄷㄱㅁㄹㅇ", "대구마리아", x)
x <- gsub("ㅅㅍㅁㄹㅇ", "송파마리아", x)
x <- gsub("ㅂㅅㅁㄹㅇ", "부산마리아", x)
x <- gsub("ㅅㅂㅁㄹㅇ", "상봉마리아", x)
x <- gsub("ㅇㅅㅁㄹㅇ", "일산마리아", x)
x <- gsub("ㄷㅈㅁㄹㅇ", "대전마리아", x)
x <- gsub("ㅅㅈㅁㄹㅇ", "수지마리아", x)
x <- gsub("ㅍㅊㅁㄹㅇ", "평촌마리아", x)
x <- gsub("ㅂㅊㅁㄹㅇ", "부천마리아", x)
x <- gsub("ㅅㅅ마리아", "신설마리아", x)
x <- gsub("ㄷㄱ마리아", "대구마리아", x)
x <- gsub("ㅅㅍ마리아", "송파마리아", x)
x <- gsub("ㅂㅅ마리아", "부산마리아", x)
x <- gsub("ㅅㅂ마리아", "상봉마리아", x)
x <- gsub("ㅇㅅ마리아", "일산마리아", x)
x <- gsub("ㄷㅈ마리아", "대전마리아", x)
x <- gsub("ㅅㅈ마리아", "수지마리아", x)
x <- gsub("ㅍㅊ마리아", "평촌마리아", x)
x <- gsub("ㅂㅊ마리아", "부천마리아", x)
x <- gsub("ㅈㅇ", "제일병원", x)
x <- gsub("ㅊㅂㅇ", "차병원", x)
x <- gsub("홍양", "생리", x)
x <- gsub("단호박", "임테기한줄", x)
x <- gsub("셤관", "시험관", x)
x <- gsub("화유", "화학적유산", x)
x <- gsub("계유", "계류유산", x)
x <- gsub("자임", "자연임신", x)
x <- gsub("숙제", "성관계", x)
x <- gsub("난저", "난소기능저하", x)
x <- gsub("신랑", "남편", x)
x <- gsub("피검", "피검사", x)
x <- gsub("직장", "회사", x)
x <- gsub("촘파", "초음파", x)
x <- gsub("[^A-Za-z가-힣[:space:][:digit:][:punct:]]", "", x)
x <- gsub("@|\n|RT", "", x)
x <- gsub("[[:punct:]]", " ", x)
x <- gsub("[[:digit:]]", "", x)
x <- gsub("인공차", "인공", x)
x <- gsub("신선차", "신선", x)
x <- gsub("피검차", "피검사", x)
x <- gsub("냉동차", "냉동", x)
x <- tolower(x)
x <- gsub("[a-z]", "", x)
x <- gsub("[\t\n]", "", x)
cafe <- cbind(date= date1, url = n_url)
cafe_gen[[length(cafe_gen)+1]] <- cafe
cafe_content[[length(cafe_content)+1]] <- x
}
}
# 클아이언트 종료 (작업 종료 후)
remDr$close()
# 서버종료
rD[["server"]]$stop()
########################################################
useNIADic()
# 명사/ 형용사/ 동사 추출
ko.words <- function(doc){
d <- as.character(doc)
pos <- paste(SimplePos09(d))
extracted <- str_match(pos, '([가-힣0-9]+)/N')
keyword <- extracted[,2]
keyword[!is.na(keyword)]
}
# SimplePos22(doc)
# extractNoun(doc)
# doc <- "회원님 점심은 도시락 준비하겠습니다"
# doc <- "회원님 점심 감사합니다"
# ko.words(doc)
# https://www.rdocumentation.org/packages/tm/versions/0.7-6/topics/VectorSource
# install.packages("tm")
library(tm)
cps <- VCorpus(VectorSource(cafe_content))
# https://www.rdocumentation.org/packages/KoNLP/versions/0.80.1/topics/SimplePos09
# https://kbig.kr/portal/kbig/datacube/niadict.page
# https://www.rdocumentation.org/packages/stringr/versions/1.4.0/topics/str_match
tdm <- TermDocumentMatrix(cps,
control = list(weighting= weightBin,
tokenize=ko.words,
removePunctuation = T,
removeNumbers = T,
stopwords = c()))
tdm <- as.matrix(tdm)
# View(tdm)
v <- sort(slam::row_sums(tdm), decreasing = T)
data <- data.frame(X=names(v),freq=v)
table(data$freq)
data1 <- data[data$freq>=10,]
# https://cran.r-project.org/web/packages/wordcloud2/vignettes/wordcloud.html
# install.packages("wordcloud2")
library(wordcloud2)
wordcloud2(data1)
wordcloud2(data1, color = "random-light", backgroundColor = "grey")
################################################################
# 키워드 데이터 불러오기
# install.packages("openxlsx")
library("openxlsx")
# xls 패키지 있는 경우에는 detach
# detach("package:xlsx", unload = TRUE)
keyword <- read.xlsx(xlsxFile="키워드수정.xlsx", sheet=1, rows = c(1:271), cols=c(1),
colNames=TRUE)
################################################################
# TM에서 keyword 존재하는 것만 남기기
freq.words <- tdm[row.names(tdm) %in% keyword$x3, ]
co.matrix <- freq.words %*% t(freq.words)
write.csv(co.matrix, "co.matrix.csv")
setwd("C:/Users/user/Desktop/단기과정/서울역차병원")
# save.image(file="moms7adj.Rdata")
# load("moms7adj.Rdata")
네트워크 분석
#################################################################################
# 패키지 설치 및 실행
# install.packages("RSelenium")
# install.packages("rvest")
# install.packages("httr")
# install.packages("stringr")
# install.packages("dplyr")
# install.packages("KoNLP")
# install.packages("RColorBrewer")
# install.packages("wordcloud")
# install.packages("tm")
# install.packages("igraph")
# install.packages("sqldf")
# install.packages("reshape")
library(KoNLP)
library(RSelenium)
library(rvest)
library(httr)
library(stringr)
library(dplyr)
library(RColorBrewer)
library(wordcloud)
library(tm)
library(igraph)
library(sqldf)
library(reshape)
setwd("C:/Users/user/Desktop/단기과정/서울역차병원")
load("moms7adj.Rdata")
options(stringsAsFactors = FALSE)
################################################################
# 키워드 데이터 불러오기
# install.packages("openxlsx")
library("openxlsx")
# xls 패키지 있는 경우에는 detach
# detach("package:xlsx", unload = TRUE)
keyword <- read.xlsx(xlsxFile="키워드수정.xlsx", sheet=1, rows = c(1:271), cols=c(1),
colNames=TRUE)
################################################################
# TDM에서 keyword 존재하는 것만 남기기
freq.words <- tdm[row.names(tdm) %in% keyword$x3, ]
############################################################################################
# KeyWord에 대한 관계 분석
tf <- apply(freq.words, 1, sum)
morp <- row.names(freq.words)
morp_tf <- data.frame(morp=morp, tf=tf)
row.names(morp_tf) <- NULL
ttfa <- morp_tf %>% summarise(ttfa=sum(tf))
morp_tf$p_a <-
as.numeric(sapply(morp_tf$tf, function(x) { round(x/ttfa*100, digits=2)} ))
morp_tf <- morp_tf %>% arrange(desc(p_a)) %>% mutate(rank = row_number())
top_keyword <- morp_tf[morp_tf$rank<=10, ]
dt_all <- data.frame(morp=morp, freq.words)
row.names(dt_all) <- NULL
dt_all1 <- melt(dt_all, id.vars="morp")
dt_all2 <- dt_all1[dt_all1$value>0, ]
dt_all2$variable <- as.numeric(gsub("X","",dt_all2$variable))
dt_all2 <- dt_all2 %>% dplyr::rename(src_keyid = variable, etf = value) %>%
arrange(src_keyid, morp)
media_anal <- function(){
morp_tf5 <- top_keyword %>% select(morp, p_a) # 상위 10개 키워드
colnames(morp_tf5)[1] <- "term"
dt_all3 <- dt_all2 # head(dt_all3, 20) # 키워드별 문서별 개수
colnames(dt_all3)[1] <- "term"
morp_tf6 <- morp_tf # head( morp_tf6 ) # 키워드별 개수
colnames(morp_tf6)[1] <- "term"
morp_tf7 <- c()
y <- 1
for (i in morp_tf5$term ){ # i <- "이식"
dt_all4 <- dt_all3[dt_all3$term ==i,]
dt_all4 <- dt_all3[dt_all3$src_keyid %in% unique(dt_all4$src_keyid),]
dt_all5 <- sqldf("
select term, count(*) as count, sum(etf) as tf from dt_all4
group by term
order by count desc
", drv = "SQLite")
vf4 <- sqldf("
select count(*) as tcount from
( select distinct src_keyid from dt_all4 )
", drv = "SQLite")
dt_all5$count <- as.numeric(dt_all5$count)
vf4$tcount <- as.numeric(vf4$tcount)
vf5 <- sqldf("
select a.*, b.tcount, a.count/b.tcount*100 as p_a_given_b from dt_all5 a, vf4 b
", drv = "SQLite")
vf8 <- vf5[vf5$p_a_given_b>=5,] # 5% 이상으로 Cut
vf8$p_a_given_b <- as.numeric(vf8$p_a_given_b)
morp_tf6$p_a <- as.numeric(morp_tf6$p_a)
vf9_1x <- morp_tf5[morp_tf5$term==i,]
vf9_1y <- sqldf("
select a.*, b.p_a as p_a,
a.p_a_given_b/b.p_a as d_lift
from vf8 a inner join morp_tf6 b
on a.term = b.term
order by p_a_given_b desc, d_lift desc
", drv = "SQLite")
vf9_1 <- sqldf("
select b.term as base, a.*, b.p_a as p_b
from vf9_1y a, vf9_1x b
", drv = "SQLite")
vf10 <- sqldf("
select *, p_a_given_b*p_b/100 as p_a_and_b,
p_a + p_b - p_a_given_b*p_b/100 as p_a_or_b
from vf9_1
", drv = "SQLite")
if(y==1) vf10 <- head(vf10[vf10$d_lift>=2,], 10)
if(y> 1) vf10 <- head(vf10[vf10$d_lift>=2 & !(vf10$term %in% morp_tf5$term[1:(y-1)] ),], 10)
# vf10 <- head(vf10[vf10$d_lift>=2,], 10)
morp_tf7 <- rbind(morp_tf7,vf10)
y <- y+1
}
morp_tf8 <- morp_tf7[morp_tf7[,1] != morp_tf7[,2],]
sum_p_b = sum( unique(data.frame(morp_tf8$base, morp_tf8$p_b))[,2] ) #### <- 수정
morp_tf8 = data.frame(morp_tf8, sum_p_b)
morp_tf8 = morp_tf8 %>% data.frame(p_b_i = morp_tf8$p_b / sum_p_b *100) # 주제확률 p_b_i
morp_tf8 = morp_tf8 %>% group_by(base) %>% mutate(sum_p_ab= sum(p_a_given_b), p_ab_i = p_a_given_b / sum_p_ab *100)
# gephi와 NodeXL로 내보내기
con <- file("result.csv")
temp <- morp_tf8[,c(1:3)]
colnames(temp) <- c("Source", "Target", "Weight")
write.csv(temp, file=con, row.names=FALSE )
# clustering
ex <- morp_tf8[,c(1,2,8)]
colnames(ex)[1] <- 'from'
colnames(ex)[2] <- 'to'
g <- graph.data.frame(ex, directed=FALSE)
# plot(g)
# https://sojungluciakim.wordpress.com/2016/03/18/r-igraph를-활용한-community-detection/
fgc <- fastgreedy.community(simplify(g)) # NodeXL과 같은방식
# fgc <- walktrap.community(g)
# fgc <- edge.betweenness.community(g)
membership(fgc)
sizes(fgc)
par(mar=c(0,0,0,0))
V(g)$size = 1*degree(g)
# plot(g, vertex.color=membership(fgc)) ## vertex.color 옵션 사용
# 주제별 클러스터 정보
cluster <- as.data.frame(cbind(fgc$names, fgc$membership))
cn <- c("names", "membership")
colnames(cluster) <- cn
map_lh_cluster <- morp_tf8 %>% select(base,term,p_b_i, p_ab_i)
map_cluster <- merge(map_lh_cluster,cluster,by.x="base",by.y="names")
map_cluster <- merge(map_cluster,cluster,by.x="term",by.y="names")
map_lh_cluster <- map_cluster[( map_cluster$membership.y == map_cluster$membership.x),]
map_lh_cluster <-map_lh_cluster[order(map_lh_cluster$ membership.x, decreasing=FALSE), ]
# 클러스터별 주요 문서
# 1) 특정 클러스터만
cluster_topic <- function(cluster, cluster_nm){ # cluster_nm = 1
tp_cluster_df = cluster %>% filter(names %in% morp_tf5$term) %>% arrange(membership)
#base
topic_nm <- as.character(tp_cluster_df[which(tp_cluster_df$membership == cluster_nm),]$names)
#term
topic_term <- (morp_tf8 %>% filter(base %in% topic_nm))$term %>% unique
#pop by base
pop_tn <- (dt_all3 %>% filter(grepl(paste(topic_nm,collapse="|"),term)))$src_keyid %>% unique
# head(pop_tn)
pop_by_base <- dt_all3 %>% filter(src_keyid %in% pop_tn)
# head(pop_by_base)
topic_tn <- c()
for(i in topic_term){ # i <- "이식"
term_cont <- pop_by_base %>% filter(term == i) %>% select(src_keyid)
topic_tn <- rbind(topic_tn,term_cont)
} #head(topic_tn)
topic_tn <- topic_tn %>% dplyr::group_by(src_keyid) %>% dplyr::summarise(count = n()) %>% dplyr::arrange(desc(count))
topicnm_top5 <- (topic_tn %>% head(n=5))$src_keyid
topicnm_print <- as.character(cafe_content[topicnm_top5])
# topicnm_print <- as.character(cafe_gen[topicnm_top5])
return(topicnm_print)
}
# 2) 전체 클러스터 내용 확인
cluster_alltopic = function(cluster){
totaltopic <- c()
tp_cluster_df = cluster %>% filter(names %in% morp_tf5$term) %>% arrange(membership)
for(i in 1:max(as.numeric(tp_cluster_df$membership))){ # i <- 1
topic_i <- cluster_topic(cluster,i)
topic_i <- data.frame(cluster=i,keyword = paste(tp_cluster_df[tp_cluster_df$membership == i,]$names,collapse=", "),
sub_key = paste(cluster[cluster$membership == i,]$names,collapse=", "), topic_i)
totaltopic <- rbind(totaltopic,topic_i)
}
return(totaltopic)
}
topic_article <- cluster_alltopic(cluster)
write.csv(topic_article,"topic_article.csv")
}
media_anal()

