Review of Active Learning via Bayesian Optimization for Materials Discovery
jupyter notebook: https://nanohub.org/resources/bayesopt
Active Learning via Bayesian Optimization for Discovery of Energy Storage Materials
1. Problem definition for Bayesian optimization
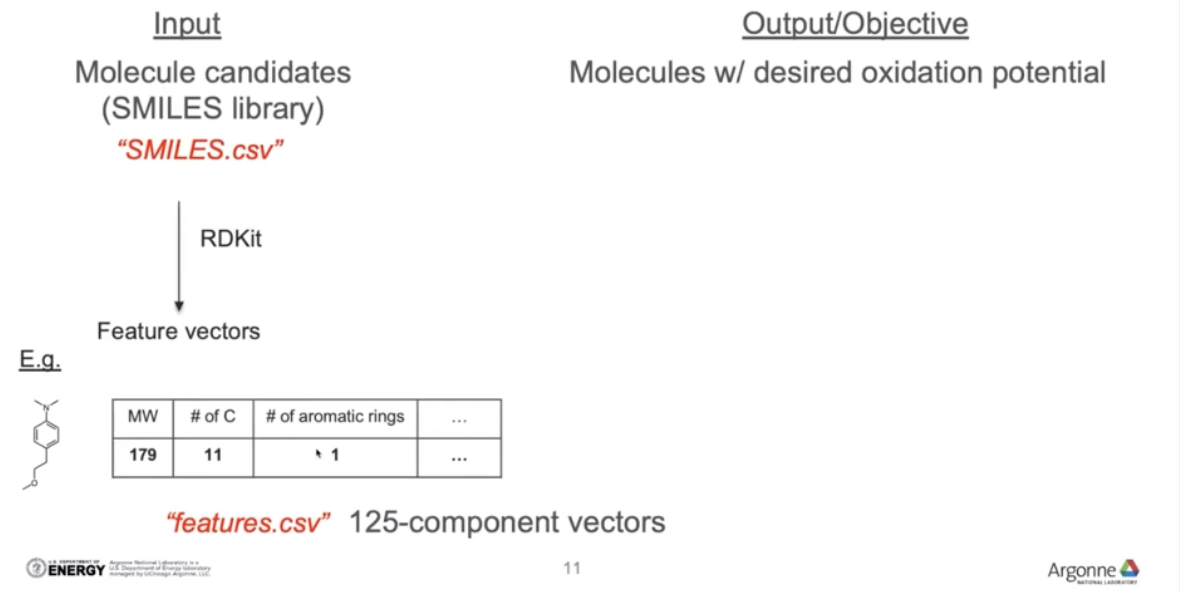
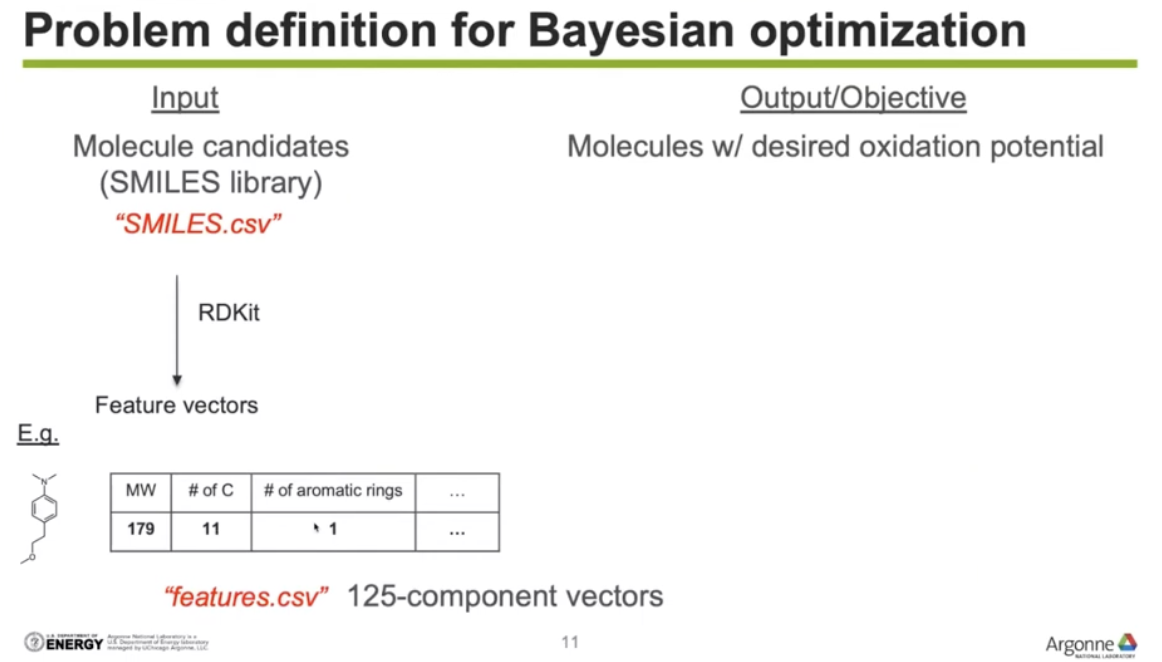
Input: Molecule candidates(SMILES library)
Output/Objective: Molecules w/ desired oxidation potential
2. Bayesian optimization scheme
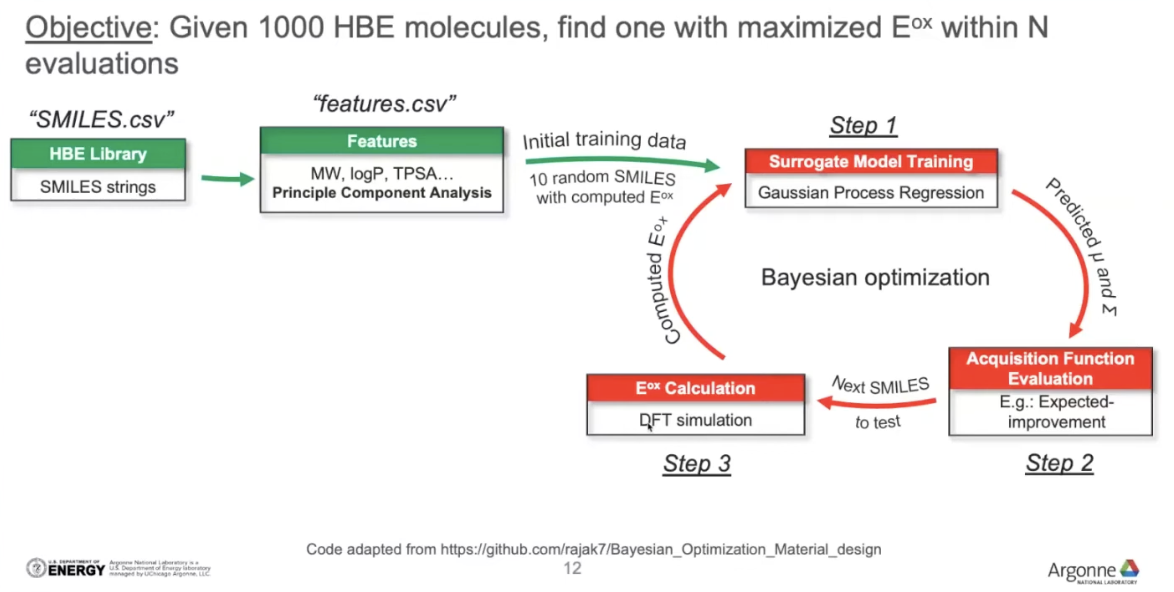
(1) SMILES string -> feature vectors(select features using PCA)
(2) [Step 1] Initialize the model
- select 10 random molecules -> train
(3) Make prediction on the remaining dataset (990 molecules)
(4) [Step 2] Acquisition Function Evaluation: determine which molecules should be evalated next(molecule that has chance to have maximum oxidation potential)
(5) [Step 3, Evaluation process] Perform DFT simulation on that particular molecule
(6) After the simulation the new molecule is made -> ADD the new molecule back to the original training set(10 molecules -> 11 molecules)
3. When to stop?
Depends on the evaluation process(Step 3)
- If you have efficient way to evaluate the property: cycle 여러 번 실행 가능
- If the evaluation process is difficult and experimental: cycle 실행 수 제한될 수 있음
Martin Fitzner "Industrial view on Bayesian optimization A perfect match for the low/no-data regime"
Patrick Rinke: Active Materials Exploration and Characterization with Bayesian Optimization
Bayesian Optimization (Bayes Opt): Easy explanation of popular hyperparameter tuning method
