SELECT COUNT(*) AS count
FROM (
SELECT id, b_v, GROUP_CONCAT(pos ORDER BY pos) AS traits
FROM (
SELECT id, b_v, pos
FROM (
SELECT id, b_v, LENGTH(b_v) - n + 1 AS pos, SUBSTRING(b_v, n, 1) AS bit_value
FROM (
SELECT id, BIN(GENOTYPE) AS b_v, t.n
FROM ECOLI_DATA
CROSS JOIN (
SELECT 1 AS n UNION ALL SELECT 2 UNION ALL SELECT 3 UNION ALL SELECT 4
UNION ALL SELECT 5 UNION ALL SELECT 6 UNION ALL SELECT 7 UNION ALL SELECT 8
UNION ALL SELECT 9 UNION ALL SELECT 10 UNION ALL SELECT 11 UNION ALL SELECT 12
UNION ALL SELECT 13 UNION ALL SELECT 14 UNION ALL SELECT 15 UNION ALL SELECT 16
) t
WHERE t.n <= LENGTH(BIN(GENOTYPE))
) AS binary_positions
) AS extracted_positions
WHERE bit_value = '1'
) AS pos_query
GROUP BY id, b_v
HAVING FIND_IN_SET(2, traits) = 0 AND (FIND_IN_SET(1, traits) > 0 OR FIND_IN_SET(3, traits) > 0)
) AS filtered_data;
- 이너쿼리를 이용해서 view 테이블처럼 index를 갖고있는 t 라는 임의의 테이블 만들기
2.주어진 테이블에는 index를 체크할수 있는 table이나 attribute가 없음 - 10진수를 2진수로 바꾸어야함 (bin 메소드)
- 2진수로 바꾼 비트값의 길이를 기준으로 index를 만들어야함 (조건문 where bit_value ='1')
- SUBSTRING(b_v, n, 1)은 왼쪽에서 오른쪽으로 n번째 비트 값을 추출 함
- LENGTH(b_v) - n + 1 은 오른쪽에서 왼쪽으로 n번째 비트 인덱스를 찾음
- cross join을 통해 n은 단일 열안에 "키"로써, 값이 변화함.즉 임시적인 파생테이블 t 안에서 1에서부터 16까지 변수값을 찾아서 돌아감 ( 즉 n은 key, iterator 역할 , t는 순차적 숫자 인덱스 생성용 테이블 )
CROSS JOIN (
SELECT 1 AS n UNION ALL SELECT 2 UNION ALL SELECT 3 UNION ALL SELECT 4
UNION ALL SELECT 5 UNION ALL SELECT 6 UNION ALL SELECT 7 UNION ALL SELECT 8
UNION ALL SELECT 9 UNION ALL SELECT 10 UNION ALL SELECT 11 UNION ALL SELECT 12
UNION ALL SELECT 13 UNION ALL SELECT 14 UNION ALL SELECT 15 UNION ALL SELECT 16
) t- UNION ALL 은 각 숫자 행을 생성하기 위한 방법 SQL에서 반복적으로 숫자 행을 생성할 때 사용
SELECT id, b_v, GROUP_CONCAT(pos ORDER BY pos) AS traits- GROUP_CONCAT로 각 ID에 대해 1인 비트의 위치를 결합하여 출력.
- (pos ORDER BY pos) 하는 traits 의 값이 오름차순을 나오게 하기 위해서임.
- traits = group_concat()으로 생성된 "1,2,3" 쉼표가 포함된 문자열.(varchar)
GROUP_CONCAT(pos ORDER BY pos)order by 안해도 실은 자동으로 오름차순 화해서 나옴.
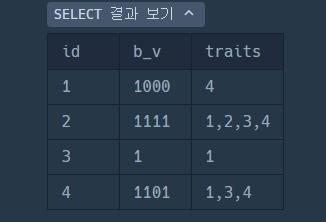
GROUP BY id, b_v
HAVING FIND_IN_SET(2, traits) = 0 AND (FIND_IN_SET(1, traits) > 0 OR FIND_IN_SET(3, traits) > 0)
) AS filtered_data;- FIND_IN_SET() 함수란 ?
MySQL에서 문자열 내의 특정 값이 있는지 확인하는 함수임.
즉,
traits 에서 "2"는 제외 (조건1)
AND
traits에서 "3" OR "1"이 포함됬는지 (조건2).
발견하지 못했을때 return 0;
syntax: FIND_IN_SET(value, string_list)
