1. 데이터준비하기
1-1 deepchem에서 데이터 불러오기(Lipophilicity of CMPD)
!wget -q "http://deepchem.io.s3-website-us-west-1.amazonaws.com/datasets/Lipophilicity.csv" -O Lipophilicity.csv
import pandas as pd
data = pd.read_csv('Lipophilicity.csv')
data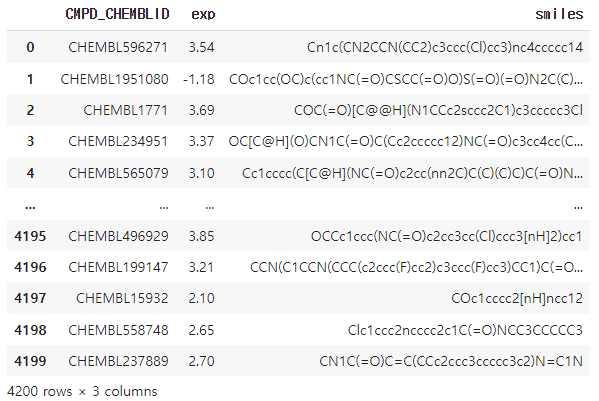
1-2 SMILES --> Molecular FingerPrint 생성
!pip install rdkit-pypi
from rdkit import Chem, DataStructs
from rdkit.Chem import AllChem
import numpy as np
fps = []
for i, smiles in enumerate(data["smiles"]):
mol = Chem.MolFromSmiles(smiles)
arr = np.zeros((1,))
fp = AllChem.GetMorganFingerprintAsBitVect(mol, 2048)
DataStructs.ConvertToNumpyArray(fp, arr)
fps.append(arr)
data["fp"] = fps
data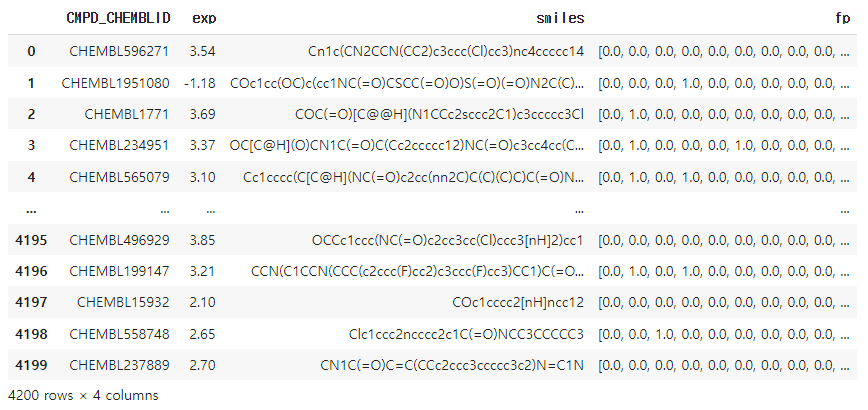
Molecule Fingerprint?
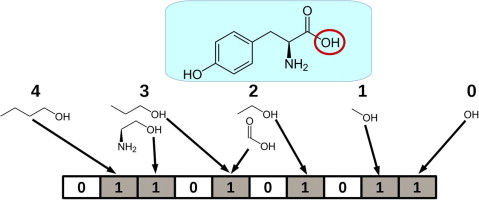 rdkit의 fingerprint 함수를 사용해서 분자의 특성 수치화
rdkit의 fingerprint 함수를 사용해서 분자의 특성 수치화
1-3 train_test_split
len(datasets['train']),len(datasets['val']),len(datasets['test'])(3359, 421, 420)
1-4 Dataset Customization [LipoDataset]
from torch.utils.data import Dataset
class LipoDataset(Dataset):
def __init__(self, df):
self.x = list(df["fp"])
self.y = list(df["exp"].values)
def __len__(self):
return len(self.x)
def __getitem__(self, index):
return torch.tensor(self.x[index]).float(), torch.tensor(self.y[index]).float()
lipodata = {
"train": LipoDataset(datasets["train"]),
"val": LipoDataset(datasets["val"]),
"test": LipoDataset(datasets["test"]),
}2. 모델구축
2-1 MLP
import torch.nn as nn
class MLP(nn.Module):
def __init__(self):
super(MLP, self).__init__()
self.linear1 = nn.Linear(2048, 1024) # 완전 연결된 단순 레이어 만들기
self.linear2 = nn.Linear(1024, 256)
self.output = nn.Linear(256, 1) # 최종 예측을 위해 출력 부 크기 1로 설정
self.dropout1 = nn.Dropout(0.5) #일반화를 위한 Dropout
self.relu = nn.ReLU()
def forward(self, x):
out = self.linear1(x)
out = self.dropout1(out)
out = self.relu(out)
out = self.linear2(out)
out = self.dropout1(out)
out = self.relu(out)
out = self.output(out)
return out
model = MLP()
model2-2 Dataloader
데이터 로더에 배치크기(1번에 입력으로 줄 단위)를 정해줄 수 있어 별도의 데이터 배치 크기 조절이 필요 없다.
from torch.utils.data import DataLoader
dataloaders = {
"train": DataLoader(lipodata["train"], batch_size=32, shuffle=True),
"val": DataLoader(lipodata["val"], batch_size=32, shuffle=False),
"test": DataLoader(lipodata["test"], batch_size=32, shuffle=False)
}
dataloaders3. 모델학습
3-1 최적화 함수 정의
import torch.optim as optim
loss_fn = nn.MSELoss() # MEAN Squared Error 회귀 지표 : MAE, MSE, RMSE, R2
optimizer = optim.Adam(model.parameters(), lr = 0.005)3-2 학습
torch.manual_seed(42)
epochs = 51
train_losses = []
val_losses = []
for epoch in range(epochs):
model.train()
epoch_train_loss = 0.0
for i, data in enumerate(dataloaders["train"]):
x, y = data[0], data[1]
optimizer.zero_grad()
outputs = model(x)
train_loss = loss_fn(outputs, y.view(-1, 1))
train_loss.backward()
optimizer.step()
epoch_train_loss += train_loss.item()
model.eval()
epoch_val_loss = 0.0
with torch.inference_mode():
for i, data in enumerate(dataloaders["val"]):
x, y = data[0], data[1]
outputs = model(x)
val_loss = loss_fn(outputs, y.view(-1, 1))
epoch_val_loss += val_loss.item()
epoch_train_loss /= len(dataloaders["train"])
epoch_val_loss /= len(dataloaders["val"])
train_losses.append(epoch_train_loss)
val_losses.append(epoch_val_loss)
if epoch % 10 == 0:
print("[Epoch %d] Train Loss: %.3f Validation Loss: %.3f" %
(epoch, epoch_train_loss, epoch_val_loss))4. 결과리포팅
import matplotlib.pyplot as plt
epoch_count = range(epochs)
plt.plot(epoch_count, train_losses, label="Train loss")
plt.plot(epoch_count, val_losses, label="Val loss")
plt.title("Training and val loss curves")
plt.ylabel("Loss")
plt.xlabel("Epochs")
plt.legend();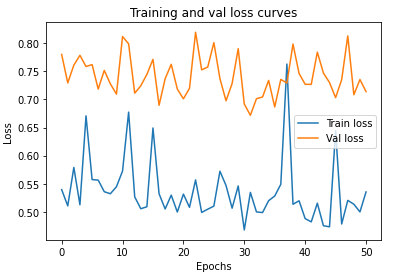
5. 훈련된 모델로 예측값 생성
from sklearn.metrics import mean_absolute_error
model.eval()
mae = 0.0
predictions = []
truths = []
with torch.inference_mode():
for data in dataloaders['test']:
x,y = data[0], data[1]
outputs = model(x)
labels = y.cpu().detach().numpy().tolist()
preds = [output[0] for output in outputs.cpu().detach().numpy().tolist()]
truths += labels
predictions += preds
mae += mean_absolute_error(labels, preds)
mae /= len(dataloaders["test"])
mae