code 출처 github : https://github.com/MIC-DKFZ/nnUNet/blob/master/nnunetv2/training/nnUNetTrainer/nnUNetTrainer.py#L271
nnUNet의 model 구조를 바꾸기 위해 정리해보는 글입니다. 필요한 부분을 작성하고 지속적으로 업데이트 할 예정입니다.
__init__(self, .....)
def __init__(self, plans: dict, configuration: str, fold: int, dataset_json: dict, unpack_dataset: bool = True,
device: torch.device = torch.device('cuda')):
self.is_ddp = dist.is_available() and dist.is_initialized()
self.local_rank = 0 if not self.is_ddp else dist.get_rank()
self.device = device
# print what device we are using
if self.is_ddp: # implicitly it's clear that we use cuda in this case
print(f"I am local rank {self.local_rank}. {device_count()} GPUs are available. The world size is "
f"{dist.get_world_size()}."
f"Setting device to {self.device}")
self.device = torch.device(type='cuda', index=self.local_rank)
else:
if self.device.type == 'cuda':
# we might want to let the user pick this but for now please pick the correct GPU with CUDA_VISIBLE_DEVICES=X
self.device = torch.device(type='cuda', index=0)
print(f"Using device: {self.device}")
# loading and saving this class for continuing from checkpoint should not happen based on pickling. This
# would also pickle the network etc. Bad, bad. Instead we just reinstantiate and then load the checkpoint we
# need. So let's save the init args
self.my_init_kwargs = {}
for k in inspect.signature(self.__init__).parameters.keys():
self.my_init_kwargs[k] = locals()[k]
### Saving all the init args into class variables for later access
self.plans_manager = PlansManager(plans)
self.configuration_manager = self.plans_manager.get_configuration(configuration)
self.configuration_name = configuration
self.dataset_json = dataset_json
self.fold = fold
self.unpack_dataset = unpack_dataset
### Setting all the folder names. We need to make sure things don't crash in case we are just running
# inference and some of the folders may not be defined!
self.preprocessed_dataset_folder_base = join(nnUNet_preprocessed, self.plans_manager.dataset_name) \
if nnUNet_preprocessed is not None else None
self.output_folder_base = join(nnUNet_results, self.plans_manager.dataset_name,
self.__class__.__name__ + '__' + self.plans_manager.plans_name + "__" + configuration) \
if nnUNet_results is not None else None
self.output_folder = join(self.output_folder_base, f'fold_{fold}')
self.preprocessed_dataset_folder = join(self.preprocessed_dataset_folder_base,
self.configuration_manager.data_identifier)
# unlike the previous nnunet folder_with_segs_from_previous_stage is now part of the plans. For now it has to
# be a different configuration in the same plans
# IMPORTANT! the mapping must be bijective, so lowres must point to fullres and vice versa (using
# "previous_stage" and "next_stage"). Otherwise it won't work!
self.is_cascaded = self.configuration_manager.previous_stage_name is not None
self.folder_with_segs_from_previous_stage = \
join(nnUNet_results, self.plans_manager.dataset_name,
self.__class__.__name__ + '__' + self.plans_manager.plans_name + "__" +
self.configuration_manager.previous_stage_name, 'predicted_next_stage', self.configuration_name) \
if self.is_cascaded else None
### Some hyperparameters for you to fiddle with
self.initial_lr = 1e-2
self.weight_decay = 3e-5
self.oversample_foreground_percent = 0.33
self.num_iterations_per_epoch = 250
self.num_val_iterations_per_epoch = 50
self.num_epochs = 1000
self.current_epoch = 0
self.enable_deep_supervision = True
### Dealing with labels/regions
self.label_manager = self.plans_manager.get_label_manager(dataset_json)
# labels can either be a list of int (regular training) or a list of tuples of int (region-based training)
# needed for predictions. We do sigmoid in case of (overlapping) regions
self.num_input_channels = None # -> self.initialize()
self.network = None # -> self.build_network_architecture()
self.optimizer = self.lr_scheduler = None # -> self.initialize
self.grad_scaler = GradScaler() if self.device.type == 'cuda' else None
self.loss = None # -> self.initialize
### Simple logging. Don't take that away from me!
# initialize log file. This is just our log for the print statements etc. Not to be confused with lightning
# logging
timestamp = datetime.now()
maybe_mkdir_p(self.output_folder)
self.log_file = join(self.output_folder, "training_log_%d_%d_%d_%02.0d_%02.0d_%02.0d.txt" %
(timestamp.year, timestamp.month, timestamp.day, timestamp.hour, timestamp.minute,
timestamp.second))
self.logger = nnUNetLogger()
### placeholders
self.dataloader_train = self.dataloader_val = None # see on_train_start
### initializing stuff for remembering things and such
self._best_ema = None
### inference things
self.inference_allowed_mirroring_axes = None # this variable is set in
# self.configure_rotation_dummyDA_mirroring_and_inital_patch_size and will be saved in checkpoints
### checkpoint saving stuff
self.save_every = 50
self.disable_checkpointing = False
## DDP batch size and oversampling can differ between workers and needs adaptation
# we need to change the batch size in DDP because we don't use any of those distributed samplers
self._set_batch_size_and_oversample()
self.was_initialized = False
self.print_to_log_file("\n#######################################################################\n"
"Please cite the following paper when using nnU-Net:\n"
"Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2021). "
"nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. "
"Nature methods, 18(2), 203-211.\n"
"#######################################################################\n",
also_print_to_console=True, add_timestamp=False)
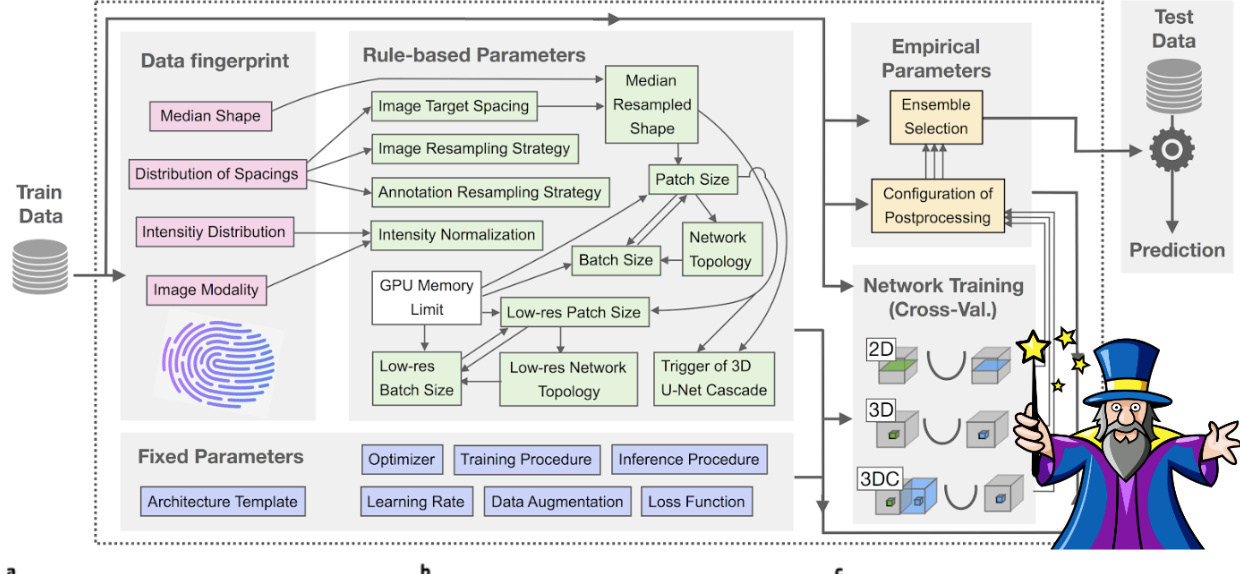
- trainer parameter 초기화하는 부분
- self.initial_lr : 초기 learning rate 지정, nnUNet 공식에 따라 lr 감소됨.
- self.num_iterations_per_epoch : epoch당 반복할 횟수 지정
- self.num_epochs : 전에 train에 진행할 epoch 지정
- self.current_epoch : 현재 epoch를 나타내는 것으로, index처럼 사용됨
- self.best_ema : 초기는 None으로, estimation이 갱신될 때 저장됨
- self.save_every : checkpoint가 저장되는 간격 지정
run_training(self)
def run_training(self):
self.on_train_start()
for epoch in range(self.current_epoch, self.num_epochs):
self.on_epoch_start()
self.on_train_epoch_start()
train_outputs = []
for batch_id in range(self.num_iterations_per_epoch):
train_outputs.append(self.train_step(next(self.dataloader_train)))
self.on_train_epoch_end(train_outputs)
with torch.no_grad():
self.on_validation_epoch_start()
val_outputs = []
for batch_id in range(self.num_val_iterations_per_epoch):
val_outputs.append(self.validation_step(next(self.dataloader_val)))
self.on_validation_epoch_end(val_outputs)
self.on_epoch_end()- 전체 train 구조를 담당
- train_epoch start 후 train_output을 저장
- validation epoch start 후 validation output 저장
- train_outputs, val_outputs : output 저장 list
on_train_start()
def on_train_start(self):
if not self.was_initialized:
self.initialize()
maybe_mkdir_p(self.output_folder)
# make sure deep supervision is on in the network
self.set_deep_supervision_enabled(self.enable_deep_supervision)
self.print_plans()
empty_cache(self.device)
# maybe unpack
if self.unpack_dataset and self.local_rank == 0:
self.print_to_log_file('unpacking dataset...')
unpack_dataset(self.preprocessed_dataset_folder, unpack_segmentation=True, overwrite_existing=False,
num_processes=max(1, round(get_allowed_n_proc_DA() // 2)))
self.print_to_log_file('unpacking done...')
if self.is_ddp:
dist.barrier()
# dataloaders must be instantiated here because they need access to the training data which may not be present
# when doing inference
self.dataloader_train, self.dataloader_val = self.get_dataloaders()
# copy plans and dataset.json so that they can be used for restoring everything we need for inference
save_json(self.plans_manager.plans, join(self.output_folder_base, 'plans.json'), sort_keys=False)
save_json(self.dataset_json, join(self.output_folder_base, 'dataset.json'), sort_keys=False)
# we don't really need the fingerprint but its still handy to have it with the others
shutil.copy(join(self.preprocessed_dataset_folder_base, 'dataset_fingerprint.json'),
join(self.output_folder_base, 'dataset_fingerprint.json'))
# produces a pdf in output folder
self.plot_network_architecture()
self._save_debug_information()
# print(f"batch size: {self.batch_size}")
# print(f"oversample: {self.oversample_foreground_percent}")
- train이 시작될 때 호출됨
- 초기화가 되지 않았다면 초기화 진행 후 output folder 생성
- 작성한 train, validation dataloader 호출
- plan을 json으로 저장 후 network_architecture plot
on_epoch_start(self)
def on_epoch_start(self):
self.logger.log('epoch_start_timestamps', time(), self.current_epoch)
- logger에 현재 timestamp 기록
on_train_epoch_start(self)
def on_train_epoch_start(self):
self.network.train()
self.lr_scheduler.step(self.current_epoch)
self.print_to_log_file('')
self.print_to_log_file(f'Epoch {self.current_epoch}')
self.print_to_log_file(
f"Current learning rate: {np.round(self.optimizer.param_groups[0]['lr'], decimals=5)}")
# lrs are the same for all workers so we don't need to gather them in case of DDP training
self.logger.log('lrs', self.optimizer.param_groups[0]['lr'], self.current_epoch)- network의 train 진행
- lr_scheduler에 의해 learning rate 조정
- logfile 출력
build_network_architecture(...)
def build_network_architecture(architecture_class_name: str,
arch_init_kwargs: dict,
arch_init_kwargs_req_import: Union[List[str], Tuple[str, ...]],
num_input_channels: int,
num_output_channels: int,
enable_deep_supervision: bool = True) -> nn.Module:
return get_network_from_plans(
architecture_class_name,
arch_init_kwargs,
arch_init_kwargs_req_import,
num_input_channels,
num_output_channels,
allow_init=True,
deep_supervision=enable_deep_supervision)
- nnunetv2/utilities/get_network_from_plans 호출
- 코드 작성자의 dynamic-network-architectures에서 필요한 network architecture를 불러와 주어진 매개변수로 모델 생성
train_step(self, ...)
def train_step(self, batch: dict) -> dict:
data = batch['data']
target = batch['target']
data = data.to(self.device, non_blocking=True)
if isinstance(target, list):
target = [i.to(self.device, non_blocking=True) for i in target]
else:
target = target.to(self.device, non_blocking=True)
self.optimizer.zero_grad(set_to_none=True)
# Autocast can be annoying
# If the device_type is 'cpu' then it's slow as heck and needs to be disabled.
# If the device_type is 'mps' then it will complain that mps is not implemented, even if enabled=False is set. Whyyyyyyy. (this is why we don't make use of enabled=False)
# So autocast will only be active if we have a cuda device.
with autocast(self.device.type, enabled=True) if self.device.type == 'cuda' else dummy_context():
output = self.network(data)
# del data
l = self.loss(output, target)
if self.grad_scaler is not None:
self.grad_scaler.scale(l).backward()
self.grad_scaler.unscale_(self.optimizer)
torch.nn.utils.clip_grad_norm_(self.network.parameters(), 12)
self.grad_scaler.step(self.optimizer)
self.grad_scaler.update()
else:
l.backward()
torch.nn.utils.clip_grad_norm_(self.network.parameters(), 12)
self.optimizer.step()
return {'loss': l.detach().cpu().numpy()}
- 실질적으로 epoch의 train을 하는 과정이 담긴 부분
- data와 target을 지정
output : network에 data를 넣어 나오는 output
- l : loss 계산 후 저장
- 이 후 backward 계산 진행
