앙상블
-
단일 머신러닝보다 좋은 성능 (정확도, 속도)
-
다수의 약한 머신러닝 모델을 종합 하여 최종 모델로 사용 --> 속도가 빠름
-
다수의 모델 구축 -> 학습데이터 분할(병렬처리) / 재사용 필요
-
단일 모델들의 취합 방법, 연결 방법에 따라
-
bagging
- with replacement (중복데이터가 하나의 train set에 들어갈 수 O)
- independent
-
boosting
- without replacement (중복되는 데이터X)
- sequential
- ( 하나의 모델 적합한 결과를 낸 후, 각각의 결과값에 가중치를 줌 -> 가중치를 받은 데이터로 또다른 모델 적합 -> 다시 가중치 줌 ) x 반복
-
K-Fold Cross Validation
- 학습 데이터를 k 개로 분할
- 검증용 분할 데이터로 순환식 사용
- 다수의 머신러닝 결과를 (통상) 평균하여 결과 도출 / 분류기는 확률적으로 결과 도출
- X_train set 중에 (모델 developing 과정에서) validation set을 뽑아, 모델 검증. (KNN)
Bootstraping
-
boosting 과 다름
-
학습 데이터로부터 n개의 학습데이터 세트 구성
-
랜덤샘플링, with replacement : 동일데이터, 동일 데이터세트에 중복 포함 가능
-
Training Data(A,B,C,D)
- Training Set1 : A,B,D,D / C : Out Of Bag (training set에 포함되지 않은 데이터 : OOB)
- Training Set2 : A,B,B,A / C,D : " "
- Training Set3 :
Bagging
- Bootstraping 으로 여러 개의 모델을 만든 다음, Aggregation (Bootstrap Aggreation)
- overfit 회피 : Weak(약한, 정확도가 낮은) 모델을 여러개 개발, 취합
Boosting
-
다수의 학습데이터 세트 구성, without replacement
-
순차적 적용 : 모델 1 개발, 개발결과를 모델 2 개발 입력값으로 활용 (병렬처리 X)
-
정확도 낮은 feature 에 대해 가중치 증가
증가된 가중치 -> 적극적인 학습 유도 (AdaBoost, Adaptive Boosting)
Gradient Tree Boosting : 순차적 모델 개발 과정에서의 가중치 조절에 Gradient Decent 사용 -
Histogram-Based Gradient Boosting : 분포도 사용, 예측값 구간화 -> 속도 향상
Voting
- 다수의 개발 모델의 취합방법 : 개별 모델의 예측값을 기초로 '투표' 실시, 최종 예측값 결정
- 개별 모델 / 예측값 별 다른 가중치 사용 가능 : score 값이 높은 모델의 투표값을 더 신뢰
Stacked
- boosting 과 비슷
- 다수의 개발 모델의 취합 방법 :개별 모델의 예측값 축적 , 최종 적용 모델의 입력값으로 사용
- 다양한 종류의 개별 모델 stacking 가능
.
.
.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import AdaBoostClassifier
from sklearn.linear_model import SGDClassifier
from sklearn.cluster import KMeans, SpectralClustering
from sklearn.metrics import r2_score, mean_squared_error
from sklearn import metricsdata = sns.load_dataset('penguins')
data.dropna(inplace = True)
data
Label Encoding
encoder = LabelEncoder()
y = encoder.fit_transform(data.iloc[:,0])MinMaxScaling
scaler = MinMaxScaler()
labels = [['bill_length_mm','bill_depth_mm','flipper_length_mm','body_mass_g']]
for i in labels :
data[i] = scaler.fit_transform(data[i])
X = data.iloc[,2:6]X_train, X_test, y_train, y_test = train_test_split(X, y, stratify = y, random_state = 10)AdaBoostClassifier
- estimator : default
m = AdaBoostClassifier()
m.fit(X_train, y_train)
print('train score', m.score(X_train, y_train)
print('test score', m.score(X_test, y_test)
# train_score 0.8112449799196787
# test_score 0.7738095238095238- estimator : SGDClassifier / n_estimators = 100
m = AdaBoostClassifier(estimator = SGDClassifier(), n_estimators=100, algorithm='SAMME')
m.fit(X_train, y_train)
print('train_score', m.score(X_train, y_train))
print('test_score', m.score(X_test, y_test))
# train_score 0.9196787148594378
# test_score 0.9404761904761905KMeans
m = KMeans(n_clusters=3, n_init = 'auto')
m.fit(X.iloc[:,:2])
print(m.labels_) # pred
# [1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
# 1 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1 1
# 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1
# 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2
# 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 2 0 2 2 2 2 2 2 2 1 2 1 2 2 0 2 2 0
# 2 2 2 2 0 2 2 2 2 2 2 0 2 2 2 1 2 0 2 2 2 2 0 0 2 1 2 2 2 0 0 0 0 0 0 0 0
# 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 2 0 0 0 1 0 0 0 0 0 0 0 0
# 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
# 0 2 0 0 0 2 0 2 0 0 0 0 0 0 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]
print(metrics.homogeneity_score(y, m.labels_))
# 0.7382437322810192sns.relplot(data, x ='bill_length_mm' , y = 'bill_depth_mm', hue= 'species')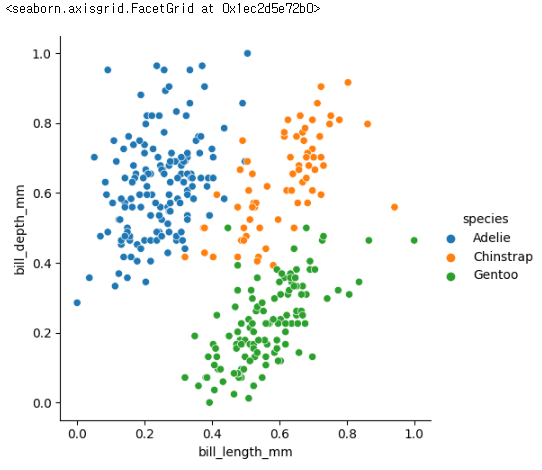
Spectral Clustering
m = SpectralClustering(n_clusters=3)
m.fit(X.iloc[:,:2])
print(m.labels_) # pred
# [2 2 2 2 2 2 2 2 2 2 2 2 1 2 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
# 2 1 2 2 2 2 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 2 2 2 2 2 2
# 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 2 2 2 2 2
# 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 1
# 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1 1 1 2 1 0 1 1 1 1 1 1 1 2 1 2 1 1 0 1 1 0
# 1 1 1 1 0 1 1 1 1 1 1 0 1 0 1 2 1 0 1 1 1 1 0 0 1 2 1 1 1 0 0 0 0 0 0 0 0
# 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 2 0 0 0 0 0 0 0 0
# 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
# 0 1 0 0 0 1 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0]
print(metrics.homogeneity_score(y, m.labels_))
# 0.6977375702902866