Khandakar Tanvir Ahmed, Jiao Sun, Sze Cheng, Jeongsik Yong, Wei Zhang
bioinformatics 2021
🐼 https://pubmed.ncbi.nlm.nih.gov/34415323/
Abstract
Motivation
-
Multi-omics data can effectively link the genotype to phenotype
-
However, the interative relation of multi-omics datasets makes it particulary challenging to incoporate different biological layers.
-
Multi-omics data integration frameworks have been proposed but, most of them ignore the relations across different biological layers in their analysis.
-
OmicsGAN, a generative adversarial network model to integrate two omics data and their integration network.
-
The model captures information from the interaction network as well as the two omics datasets and fuse them to generate synthetic data with better predictive signals.
Materials and methods
Overview of the framework
-
Input : Any two omics data with bilogical relations between each other.
-
Output : New feature sets corresponding to each omics data that contain informaion from both modality and thier interaction network.
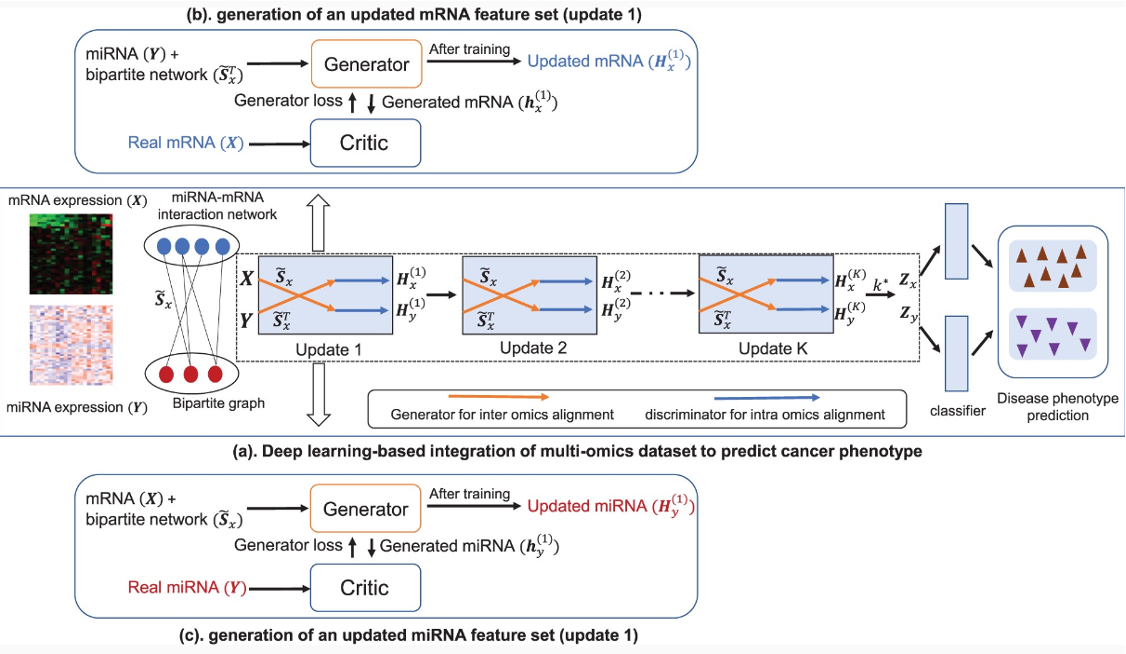
-
두 omics data 와 그들의 interaction network가 입력으로 들어간다.
-
X를 만들기 위해서 Y와 interaction network 를 사용하고, Y를 만들기 위해서는 X 와 interaction network 를 사용한다.
-
Update 를 k번 반복하며 최종 Synthetic data(, )를 만들고 classifier에 넣어 prediction을 수행한다.
-
GAN with one omics data from one distribution as input to the generator and another omics data with different distribution as real dataset in the discriminator to generate a synthetic data retaining information from both omics datasets.
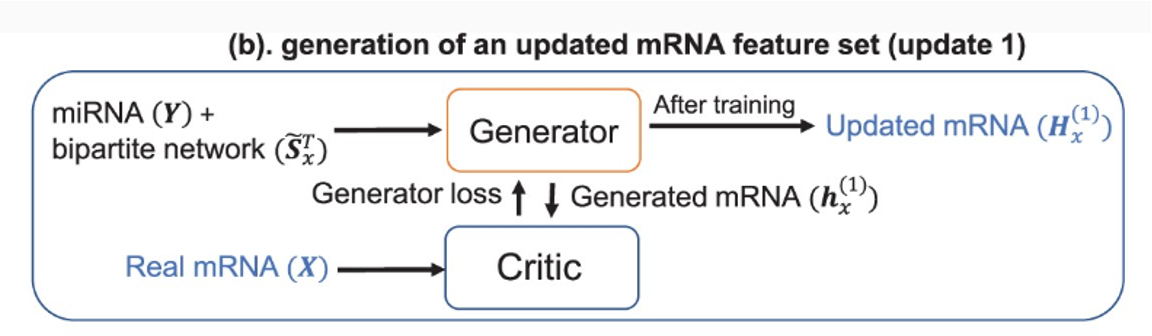
-
(b) generation of an updated mRNA feature set (update1), 하나의 omics data(miRNA)와 두 omics의 interaction network인 bipartite network()가 genetator로 들어간다. 논문에서는 discriminator 역할을 하는 Critic을 정의하였고 Critic에서 real mRNA와 generated mRNA() 간의 loss를 계산한다.그 후 Updated mRNA()가 생성된다. (Synthetic data)
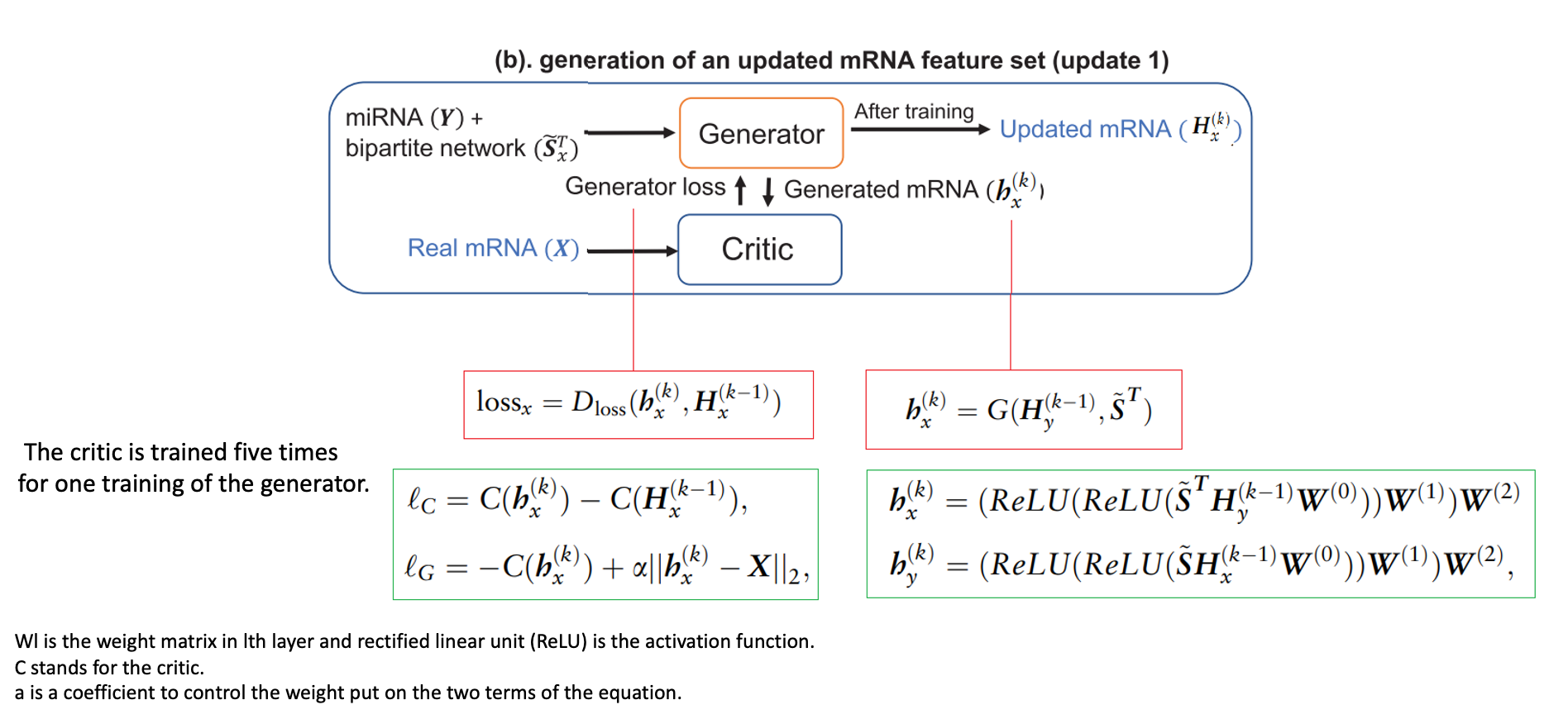
(b) with loss function
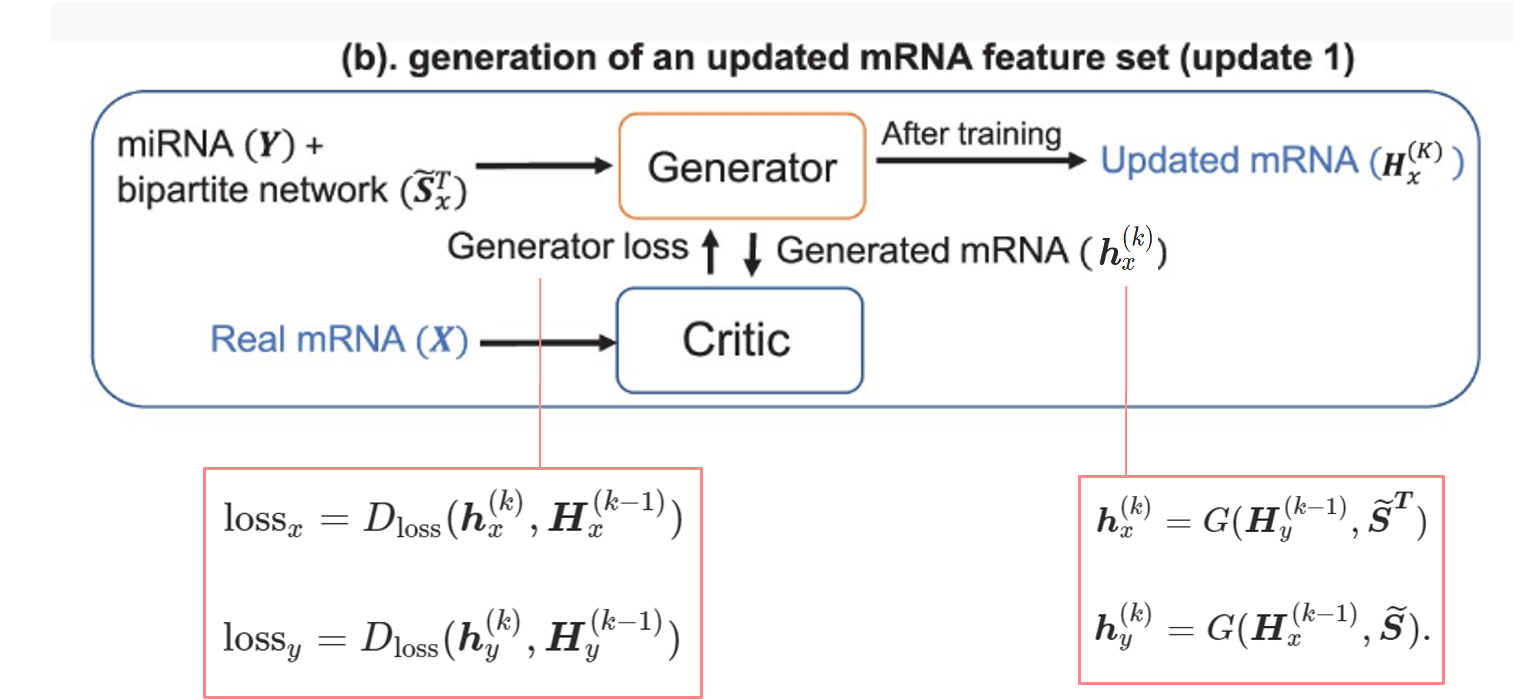
- Our proposed pipeline has two separate wGANs for two omics data to update them into a new representation. Generators in each wGAN are three layers fully connected neural network that generates a dataset based on one omics data and the normalized adjacency matrix following the equations.
Evaluation methods
- Classification model : Support vector machine(SVM) with linear kernel is implemented as a classifier for all experiments.
Trian : Test : Validation = 60% : 20% : 20% - Survival prediction model : A Cox proportional hazards model with Elastic Net penalty is applied to study the correlation between patient's overall survival and omics profiles.
Trian : Test = 80% : 20%
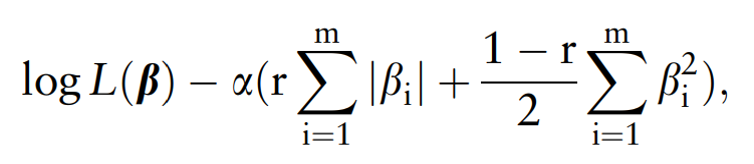
- 5-fold cross validation is performed on training data to tune the hyper-parameter α.
- α allows the model to perform feature selection while retaininf the regression coefficients of some features.
Experiments
Datasets and networks
- OmicsGAN was tested on
- TCGA breast invasive carcinoma (BRCA)- Lung adenocarcinoma (LUAD)
- Ovarian serous cystadenocarcinoma (OV) datasets
- The RNA-seq mRNA expression and miRNA expression datasets of each cancer type were downloaded from UCSC Xena Hub.
- For the mRNA expression, the log2(x+1) transformed RSEM normalized count with 20,531 genes and miRNA expression, the log2(x+1) transformed RPM value with 2,166 miRNAs.
- The miRNA-mRNA interaction network was obtained from TargetScanHuman.
- The miRNA-mRNA bipartite network contained 163,568 interaction in total (interaction was valued as -1, no interaction was valued as 1).
- In breast cancer study, we classify the cancer patients based on estrogen receptor (ER+ versus ER-) and triplet negative (TN+ versus TN-) status.
- For lung cancer and overian cancer studies, we classify the patient based on their survival time.
Running omicsGAN on the TCGA datasets
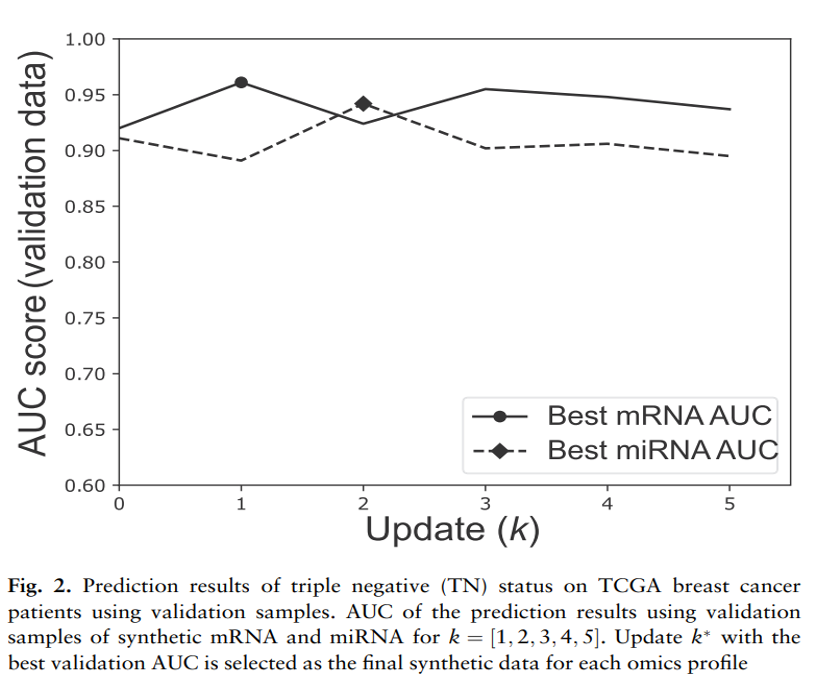
- To evaluate the proposed generation model on the TCGA omics dataset, first selection the dataset by updating the mRNA and miRNA (or TF and its target genes) expression profiles 5 times.
- One synthetic data is generated for breast cancer ER and TN status prediction based on the average validation AUC of the two clinical variables.
- 5번 update 중 validation에서 가장 높은 AUC를 가진 set (synthetic data) -> test 진행
Assumptions
- The synthetic datasets learned in omicsGAN consider the expression in both mRNA and miRNA profiles ans the violofical interacions between them. So the will provide better predictive signatures compared to mRNA and miRNA expressions.
- The better predictiva signatrues will imporve the disease phenotype prediction.
OmicsGAN improved cancer outcome prediction
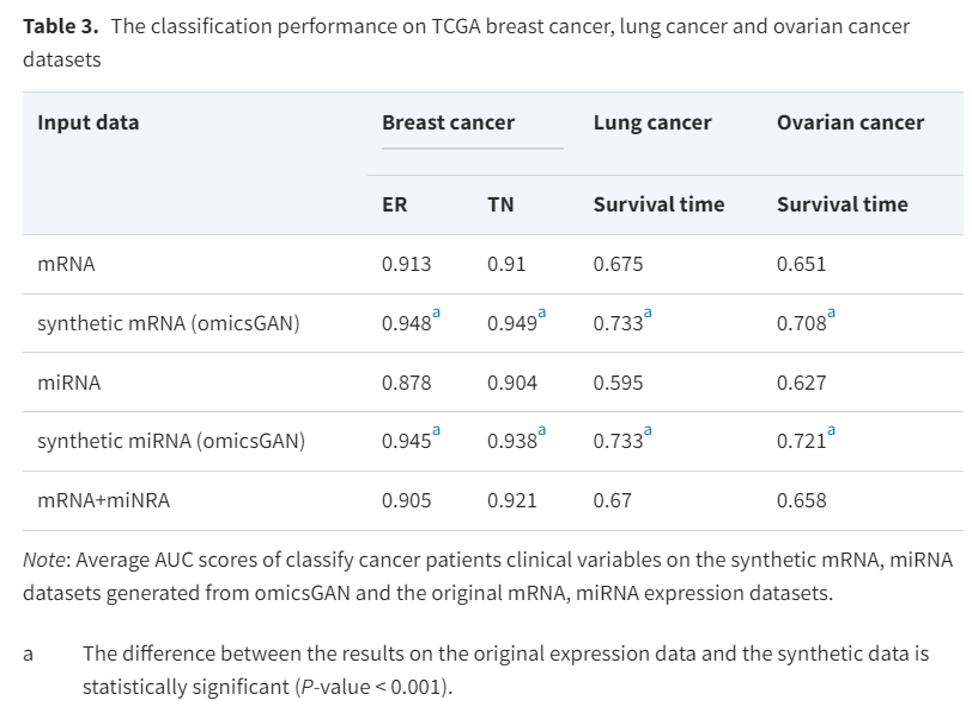
-> OmicsGAN relies in the interaciton network to generate syntheric data with better predictive signal.
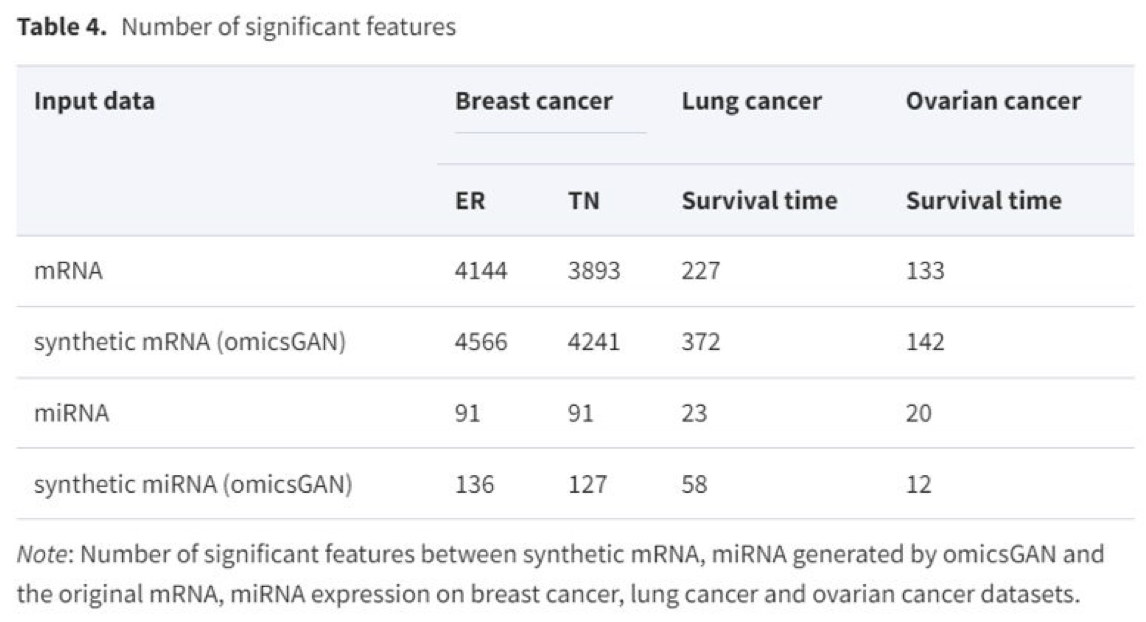
-> OmicsGAN enriches the features of synthetic datasets with better predictive singatures that results into improved cancer outcome prediction.
impaction of interaction network on cancer outcome prediction
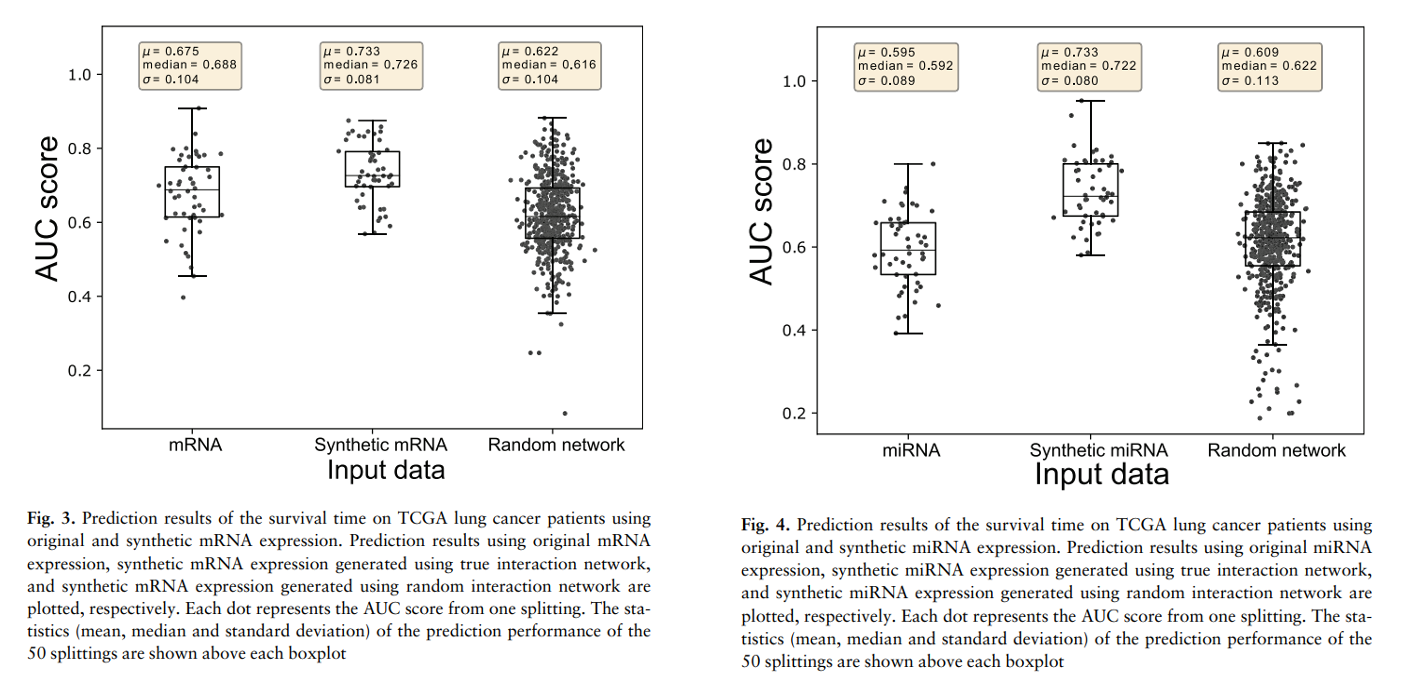
- We want to investigate whether the improvement in performance is because of the additional omics data or the model can exploit the interaction network for data integration.
- It signifies that omicsGAN does not fuse information from the two omics data directly, rather functionally incoporate the interaction network into the integration.
OmicsGAN improved survival prediction
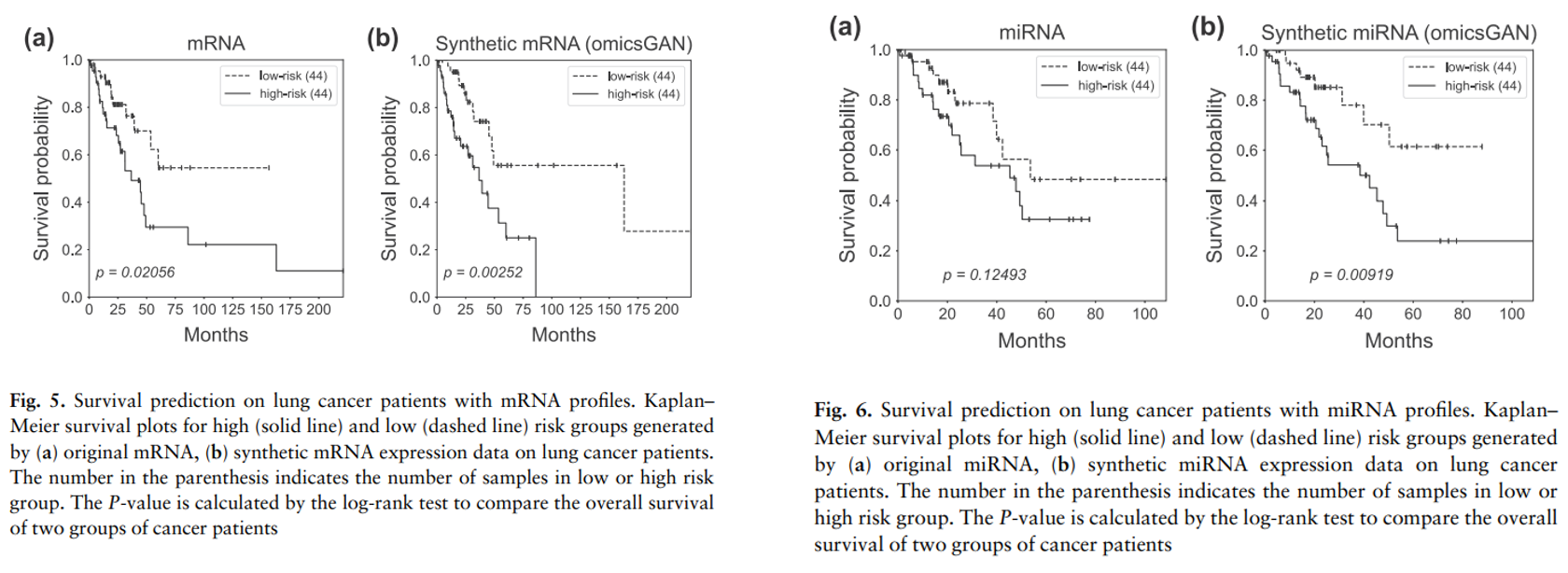
-> The log-rank test P-values clearly demonstrate a strong additional prognostic power of the synthetic omics profiles beyond the original signatures.
Integration of TF and gene expression
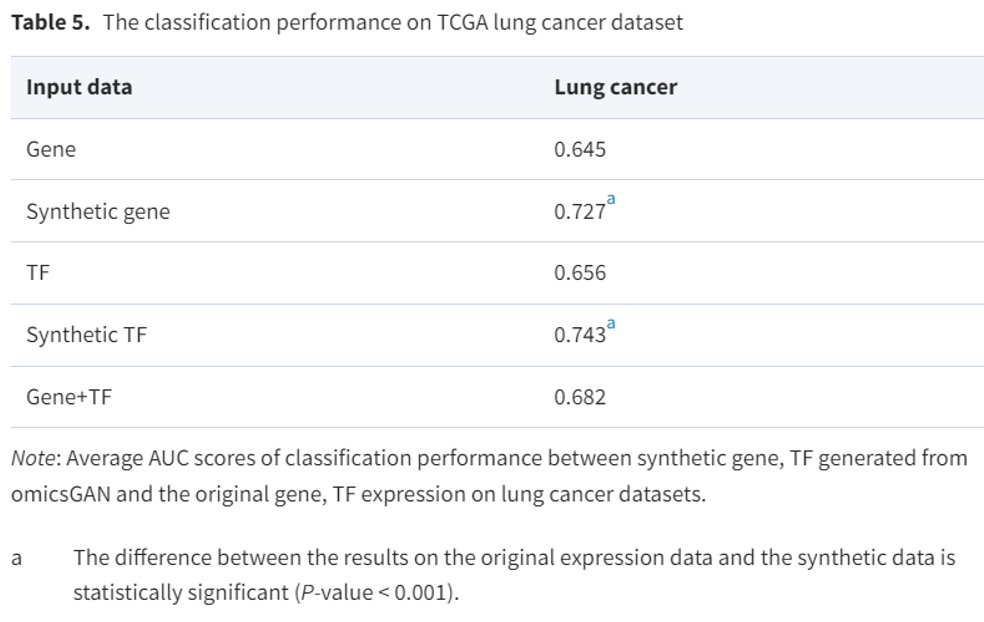
- We design another experiment using TF-gene interaction network to evaluate.
-> These finding signify that our proposed framework can work with varying set multi-omics data.
Conclusion
- Introduced omicsGAN, a GAN model to effectively integrate the interaction network and the omics datasets into new synthetic data with better predictive singals.
- We observed that the synthetic data generated from omicsGAN has better discriminative power on cancer outcome classsification and cancer patuents survival prediction compared to the original omics datasets.
- Synthetic datasets also contain more significant features that result in better predicdtive performance.
-Further experimentation show that omicsGAN does not only gather information from two omics datasets; rather funtionally incoporate their biological interaction into the integration.

