Previous Lectures
Message Passing Neural Nets
- Node features are updated from iteration t to t+1 via learnable permutation invariant neighborhood aggregate AGG and update UPD
Graph Neural Networks
- Message passing updates node features using local aggregation
- stacking multiple graph neural network - expand perceptual fields...
Normal Graph Neural Networks
- Advanced GNN layers make pooling over node features, which are then used to make a graph-level prediction
Molecular Graphs
- Molecules can be represented as a graph G with node features and edge feature
- Node features: atom type, atom charges- Edge features: valuence bond type
- However,sometimes, we also know the 3D positions , which is actually more informative
3D structure에 Graph를 어떻게 적용할 것인가?
small molecules, proteins, DNA/RNA, Inorganic Crystals, Catalysis Systems, Transportation & Logistics, Robotic Navigation ...
Geometric Graphs
- A geometric graph G = (A, S, X) is a graph where each node is embedded in d-dimensional Euclidean space.
- A: Adjacency matrix- S: scalar features
- X: tensor features
Broad Impact on Sciences
- Supervised Learning: Prediction
- Properties predictions
input: Geometric Graph → Geometric GNN → Output: Prediction - Supervised Learning: Structured Prediction
- Molecular Simulation
input: current state → Geometric GNN + Dynamics Simulator → next state → ...
분자의 안정 상태가 무엇인지를 simulate 하고 싶음. 어떻게 움직여야 하는지를 조금씩 움직여가면서 구한다. - Generative Models
- Drug or material design
input: Geometric Graph → Geometric GNN + Generative Model → Geometric Graph
What's the obstacle?
- To describe geometric graphs we use coordinate systems
- (1) and (2) use different coordinate systems to describe the same molecular geometry - We can describe the transform between coordinate systems with symmetries of Euclidean space
- 3D rotations, translations - However, ouput of traditional GNNs given (1) and (2) as completely different!
Symmetry of Inputs
- We want our GNNs can see (1) and (2) as the same systems though described differently...
- i.e., we want design Geometric GNNs aware of symmetry!
Symmetry of Outputs
- Beyond input space, output can also be tensors
- Example: simulation (force prediction)
- Given a molecule and a rotated copy, predicted forces should be the same up to rotation- i.e., Predicted forces are equivariant to rotation
Equivariance
- Formal definition of Equivariance:
a function F: X→Y is equivariant if for a transformation p is satisfies:
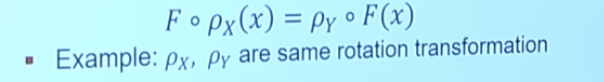
Illustration: 3D Rotation Equivariance
구조가 회전하면 힘 역시 같은 형식으로 변경되어야 한다.
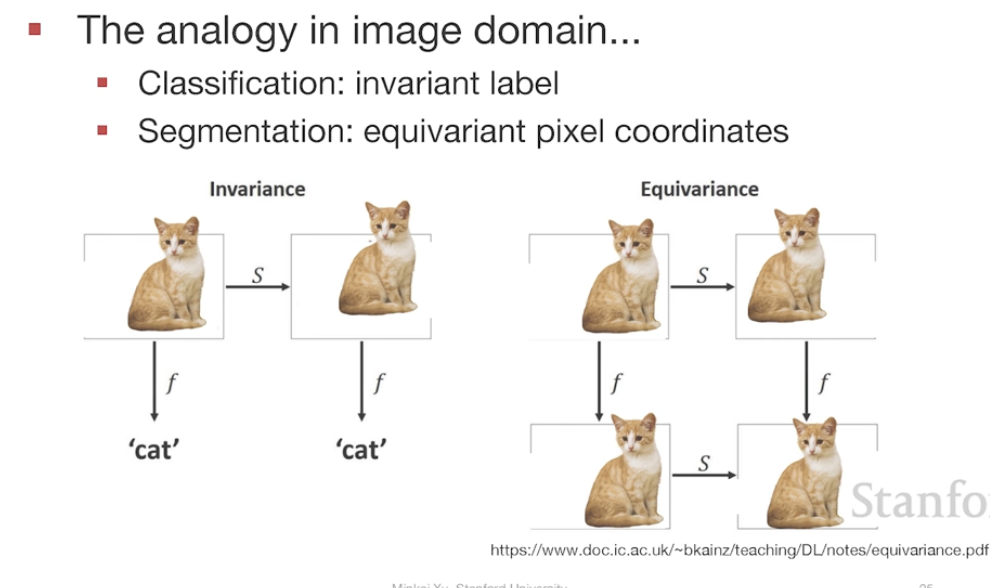
Invariance
- Definition of Invariance:
a function F:X→Y is invariant if for a transformation p it satisfies:
F * P_x(x) = F(x)
어떤 회전을 하더라도 같은 예측을 해야 한다.
Equivariance의 special case임
Invariance & Equivariance
- For geometric graphs, we consider 3D Special Euclidean (SE(3)) symmetries
- Structure x → energy E: invariant scalars
- Structure x → force v: equivalent tensors
rotation equivariant and translation invariant
Geometric GNNs
Two classes of Geometric GNNs:
- Invariant GNNs for learning invariant scalar features
- Equivariant GNNs for learning equivalent tensor features
Molecular Dynamics Simulations
- For simulating the stable structure of molecular geometrics: computiationally costly quantum mechanical calculations
For ML Models...
atom types, positions를 바탕으로 energy나 forces를 예측하고 싶음
Invariant GNNs: SchNet
- SchNet updates the node embeddings at the lth layer by message passing layers
- Schnet makes W invariant by scalarizing relative positions with relative distances
- Stack multiple interaction and atom-wise layers
- Predict single scalar value for each atom
- Sum all scalars together as energy prediction
Improved SchNet: DimeNet
- Chemcially, potential energy can be modeled as sum of four parts
- DimeNet resolves this problems by
- Do message interaction based on- Distance between atoms
- angle between bonds
Expressiveness
- Distances/Angles are imcomplete descriptors for uniquely identifying geometric structure
- This pair of geometric graphs cannot be distinguished by identical scalr quantities
- But they can be distinguished based on directional or geometric information
Equivariant GNNs: PaiNN
- PaiNN still take learnable weights W conditioned on the relative distance to control message passing
- However, differently, in PaiNN each node has two features (both scalar features and vector feature)
Molecular Conformation Generation
- Generate stable confroamtions from molecular graph
Challenges
- Generative models learns the data distribution
- Similar to the learning algorithm, geneartion process should also capture the physical symmetry groups., i.e., equiariant to roto-translation
- Diffusion model 사용 → Geometric Diffusion
