🏆 지도학습 알고리즘
분류형 선형 모델
- 특정 계수와 절편의 조합이 훈련 데이터에 얼마나 잘 맞는지 측정
- 사용할 수 있는 규제가 있는지, 있다면 ---어떤방식인지
- 그러나 알고리즘들이 만드는 잘못된 분류의 수를 최소화하도록 w와 b를 조정하는 것은 불가능
y-hat : 예측한 값
y : 정답
선형 분류 모델의 C(규제) 설정에 따른 결정 경계
import mglearn
import matplotlib.pyplot as plt
from sklearn.linear_model import LogisticRegression
from sklearn.svm import LinearSVC
import warnings
warnings.filterwarnings('ignore')
plt.rc('font', family ='NanumBarunGothic')
plt.rcParams['axes.unicode_minus'] = False
plt.rcParams['figure.dpi'] = 100
X, y = mglearn.datasets.make_forge()
fig, axes = plt.subplots(1, 2, figsize=(10, 3))
for model, ax in zip([LinearSVC(max_iter=5000), LogisticRegression()], axes):
clf = model.fit(X, y)
mglearn.plots.plot_2d_separator(clf, X, fill=False, eps=0.5,
ax=ax, alpha=.7)
mglearn.discrete_scatter(X[:, 0], X[:, 1], y, ax=ax)
ax.set_title(clf.__class__.__name__)
ax.set_xlabel("특성 0")
ax.set_ylabel("특성 1")
axes[0].legend()
plt.show()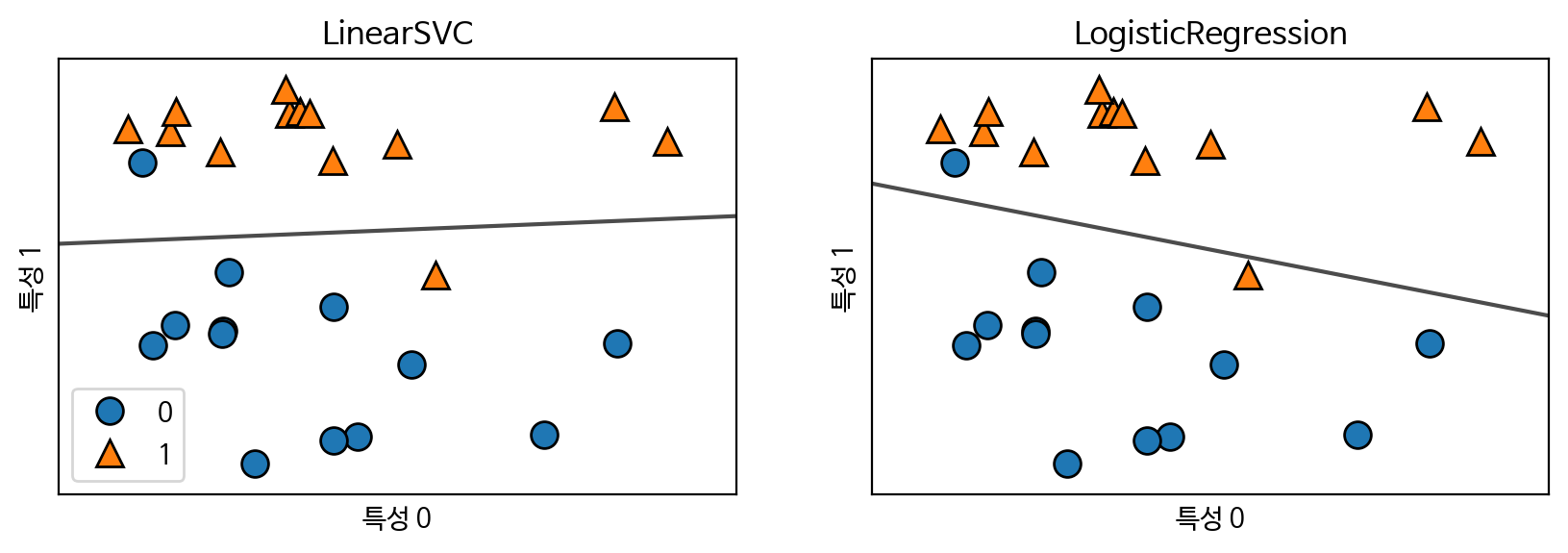
C=1 규제(공부를 덜 시키겠다 -> 과대적합을 피하겠다.) 설정 값
C 설정 값을 낮게하면 ex) 0.01, 0.001 -> 규제강화 -> 일반화 -> 과소적합
C 설정 값을 높게하면 ex) 10, 100, 1000 -> 규제완화 -> 과대적합
mglearn.plots.plot_linear_svc_regularization()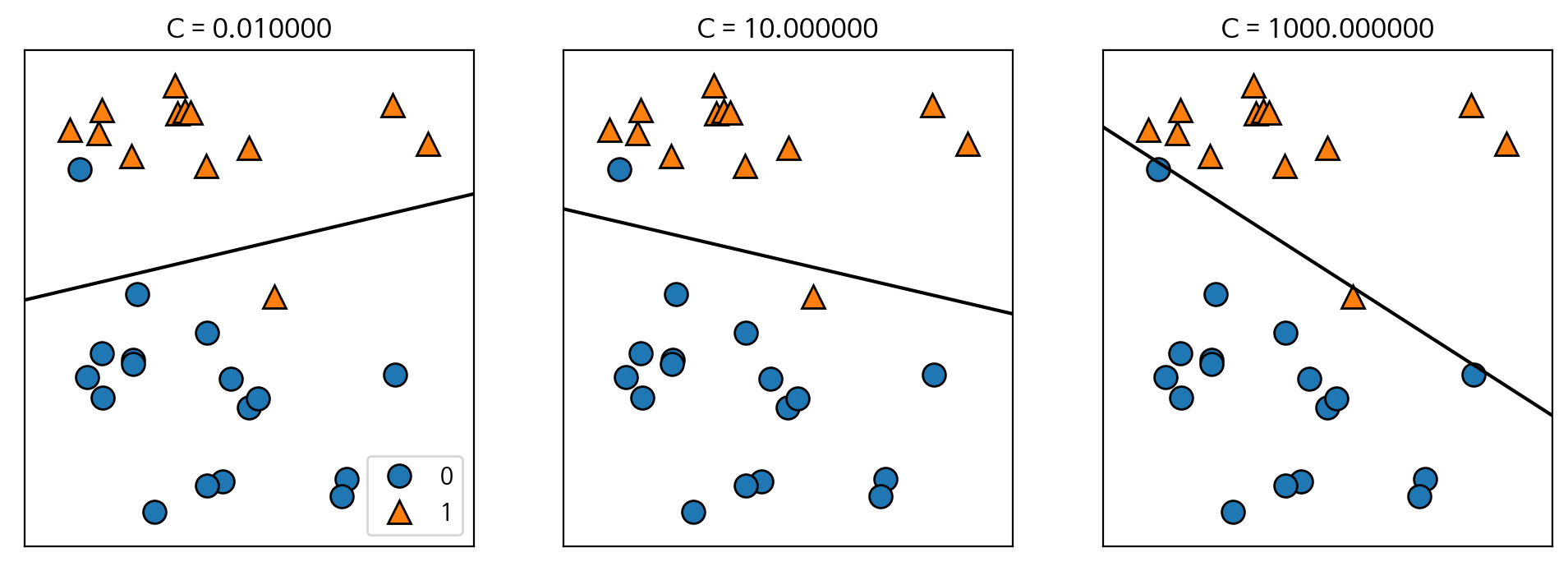
🗒️ 유방암 데이터셋을 사용한 로지스틱 회귀(LogisticRegression) 성능평가
- 로지스틱 회귀 : L2, L1모두 가능(기본은 L2)
- 규제 강도를 결정하는 C 설정에 따른 성능 비교
- 기본 C=1, ex) 규제 강화 C = 0.01, 규제 완화 C = 100
데이터 준비하기
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
# 데이터 가져오기
cancer = load_breast_cancer()
# 데이터셋 분리하기
X_train, X_test, y_train, y_test = train_test_split(cancer.data, cancer.target, stratify=cancer.target, random_state=7)👀 stratify=cancer.target : 비율 보정
모델 설정, 학습
from sklearn.linear_model import LogisticRegression
# C = 1
logreg = LogisticRegression()
logreg.fit(X_train, y_train)LogisticRegression C에 따른 규제 L2 모델 성능평가
print('-----------기본----------')
logreg001 = LogisticRegression().fit(X_train, y_train)
print('logreg 훈련 세트 점수 : {:.8f}'.format(logreg.score(X_train, y_train)))
print('logreg 테스트 세트 점수 : {:.8f}'.format(logreg.score(X_test, y_test)))
print('--------규제 강화--------')
logreg001 = LogisticRegression(C=0.01).fit(X_train, y_train)
print('logreg001 훈련 세트 점수 : {:.8f}'.format(logreg001.score(X_train, y_train)))
print('logreg001 테스트 세트 점수 : {:.8f}'.format(logreg001.score(X_test, y_test)))
print('--------규제 완화--------')
logreg100 = LogisticRegression(C=100).fit(X_train, y_train)
print('logreg100 훈련 세트 점수 : {:.8f}'.format(logreg100.score(X_train, y_train)))
print('logreg100 테스트 세트 점수 : {:.8f}'.format(logreg100.score(X_test, y_test)))👀 이 결과를 본다면, 규제 완화된 모델을 선택하는것이 좋음!
# L2 규제에 대한 feature들의 가중치를 확인
plt.plot(logreg100.coef_.T, '^', label="C=100")
plt.plot(logreg.coef_.T, 'o', label="C=1")
plt.plot(logreg001.coef_.T, 'v', label="C=0.01")
plt.xticks(range(cancer.data.shape[1]), cancer.feature_names, rotation=90)
xlims = plt.xlim()
plt.hlines(0, xlims[0], xlims[1])
plt.xlim(xlims)
plt.ylim(-5, 5)
plt.xlabel("특성")
plt.ylabel("계수 크기")
plt.legend()
plt.show()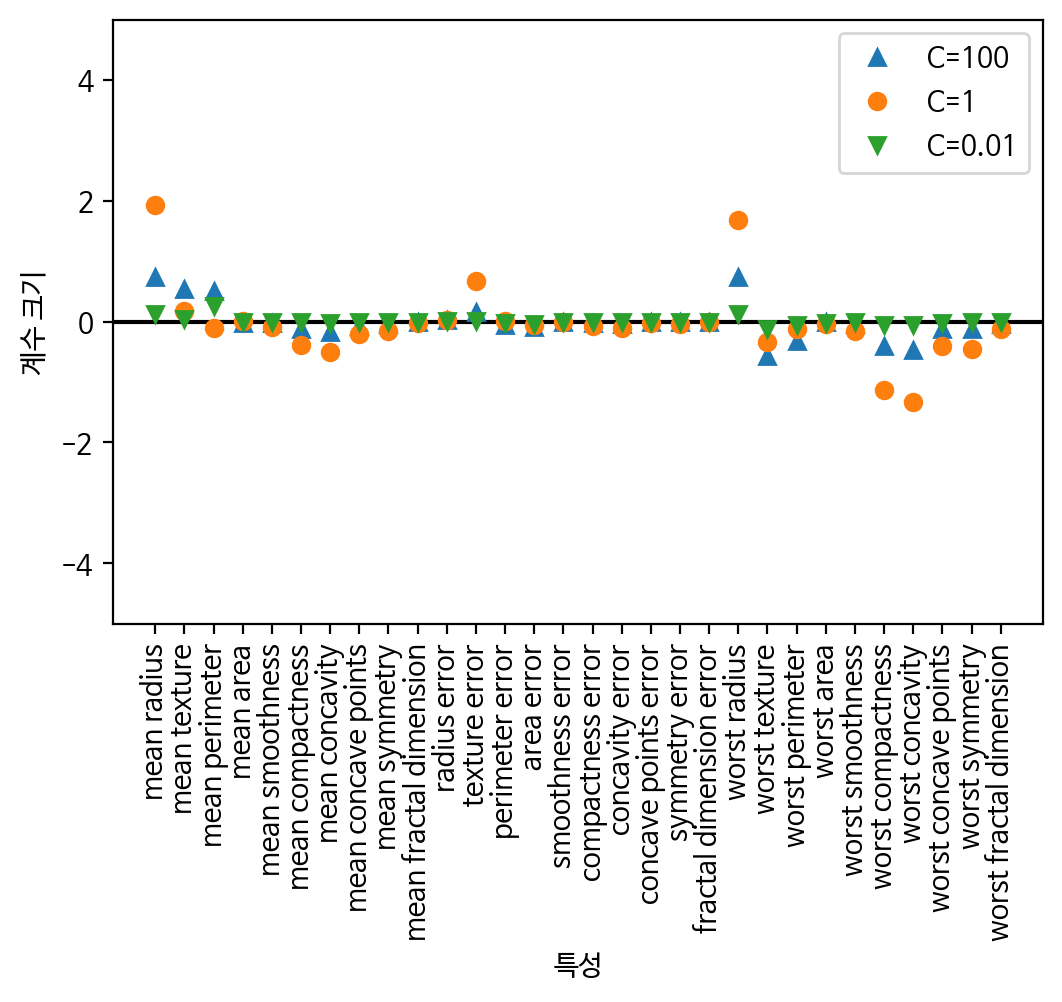
LogisticRegression C에 따른 규제 L1 모델 성능평가
print('----------기본-----------')
logreg001 = LogisticRegression(penalty='l1', solver='liblinear').fit(X_train, y_train)
print('logreg 훈련 세트 점수 : {:.8f}'.format(logreg.score(X_train, y_train)))
print('logreg 테스트 세트 점수 : {:.8f}'.format(logreg.score(X_test, y_test)))
print('--------규제 강화--------')
logreg001 = LogisticRegression(C=0.01, penalty='l1', solver='liblinear').fit(X_train, y_train)
print('logreg001 훈련 세트 점수 : {:.8f}'.format(logreg001.score(X_train, y_train)))
print('logreg001 테스트 세트 점수 : {:.8f}'.format(logreg001.score(X_test, y_test)))
print('--------규제 완화--------')
logreg100 = LogisticRegression(C=100, penalty='l1', solver='liblinear').fit(X_train, y_train)
print('logreg100 훈련 세트 점수 : {:.8f}'.format(logreg100.score(X_train, y_train)))
print('logreg100 테스트 세트 점수 : {:.8f}'.format(logreg100.score(X_test, y_test)))# L1 규제에 대한 feature들의 가중치를 확인
plt.plot(logreg100.coef_.T, '^', label="C=100")
plt.plot(logreg.coef_.T, 'o', label="C=1")
plt.plot(logreg001.coef_.T, 'v', label="C=0.01")
plt.xticks(range(cancer.data.shape[1]), cancer.feature_names, rotation=90)
xlims = plt.xlim()
plt.hlines(0, xlims[0], xlims[1])
plt.xlim(xlims)
plt.ylim(-5, 5)
plt.xlabel("특성")
plt.ylabel("계수 크기")
plt.legend()
plt.show()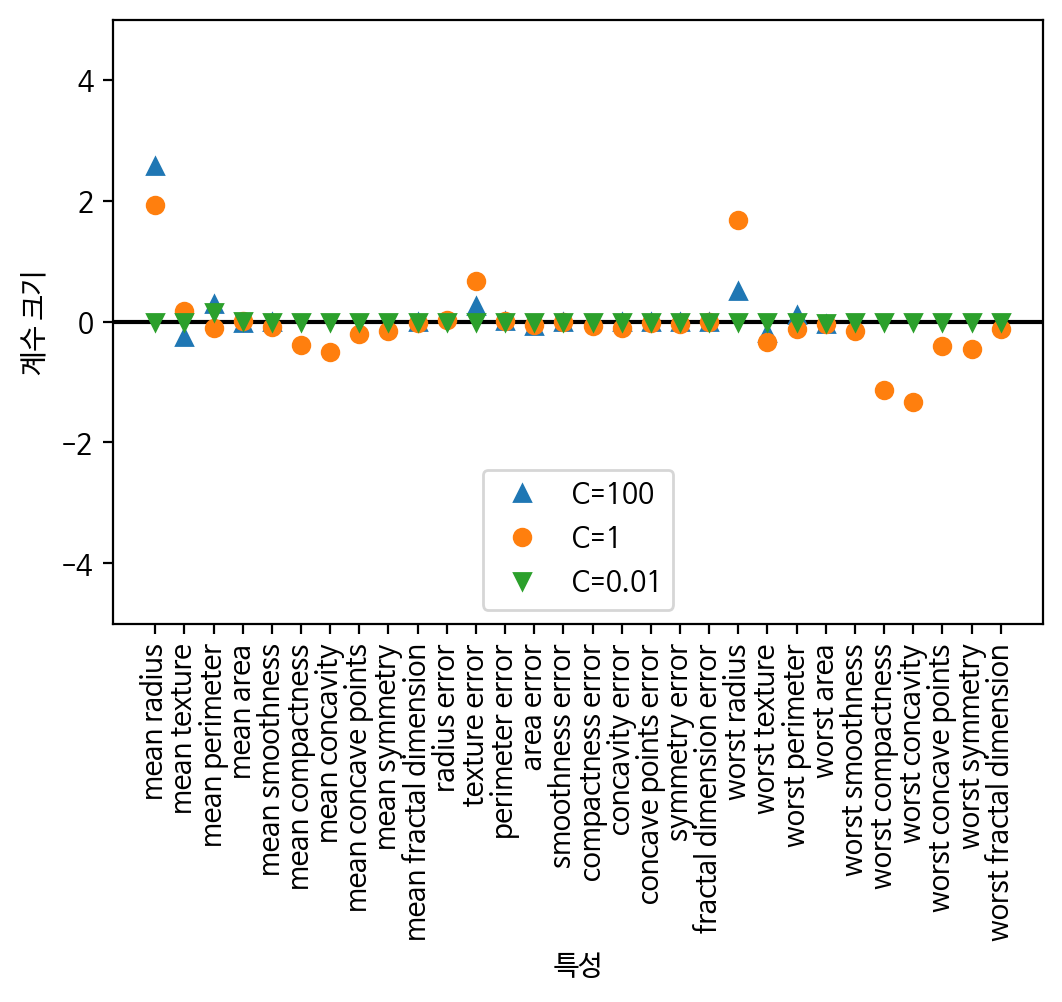
🏆 서포트 벡터 머신(SVM)
- 데이터셋의 여러 속성을 나타내는 데이터프레임의 각 열은 열 벡터 형태로 구현됨
- 열 벡터들이 각각 고유의 축을 갖는 벡터 공간 만듦 -> 분석 대상이 되는 개별 관측값은 모든 속성(열벡터) 관한 값을 해당 축의 좌표로 표시하여 벡터 공간에서 위치를 나타냄
- 속성이 2개 존재하는 데이터셋 - 2차원 평면 공간 좌표, 3개이면 3차원, 4개이면 4차원
- 벡터 공간에 위치한 훈련 데이터의 좌표와 각 데이터가 어떤 분류 값을 가져야하는지 정답을 입력받아 학습 -> 같은 분류 값을 갖는 데이터끼리 같은 공간에 위치하도록 함
🚣♀️ SVM(Support Vector Machine) 타이타닉 생존자 예측
🐼 준비
문제정의 : SVM 사용하여 타이타닉 생존자(1), 사망자(0) 예측하는 이진분류모델로 정의
라이브러리 임포트
import pandas as pd import seaborn as sns import matplotlib.pyplot as plt한글깨짐 방지
import matplotlib as mpl import matplotlib.pyplot as plt %config InlineBackend.figure_format = 'retina' !apt -qq -y install fonts-nanum import matplotlib.font_manager as fm fontpath = '/usr/share/fonts/truetype/nanum/NanumBarunGothic.ttf' font = fm.FontProperties(fname=fontpath, size=9) plt.rc('font', family='NanumBarunGothic') mpl.font_manager._rebuild()
데이터 준비하고 확인하기
# 데이터 준비하기
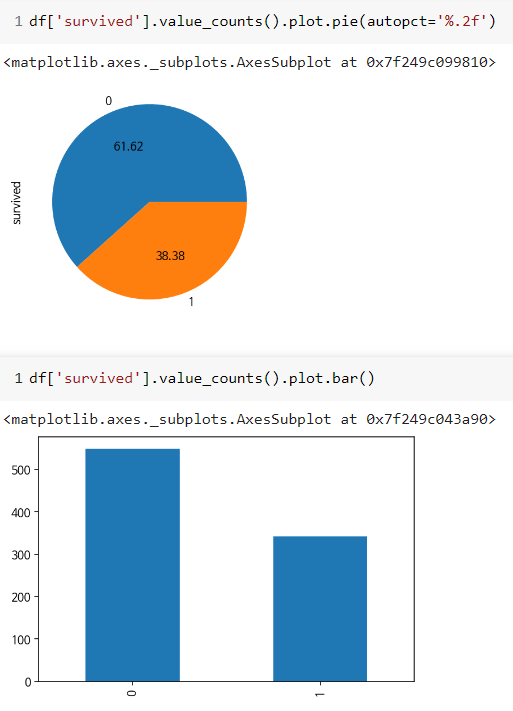
df = sns.load_dataset("titanic")
# 데이터 확인하기
df.head()
데이터 분석하기 
attrs = df.columns
plt.figure(figsize=(20,20), dpi=200)
for i, feature in enumerate(attrs):
plt.subplot(5, 5, i+1)
sns.countplot(data=df, x=feature, hue='survived')
sns.despine()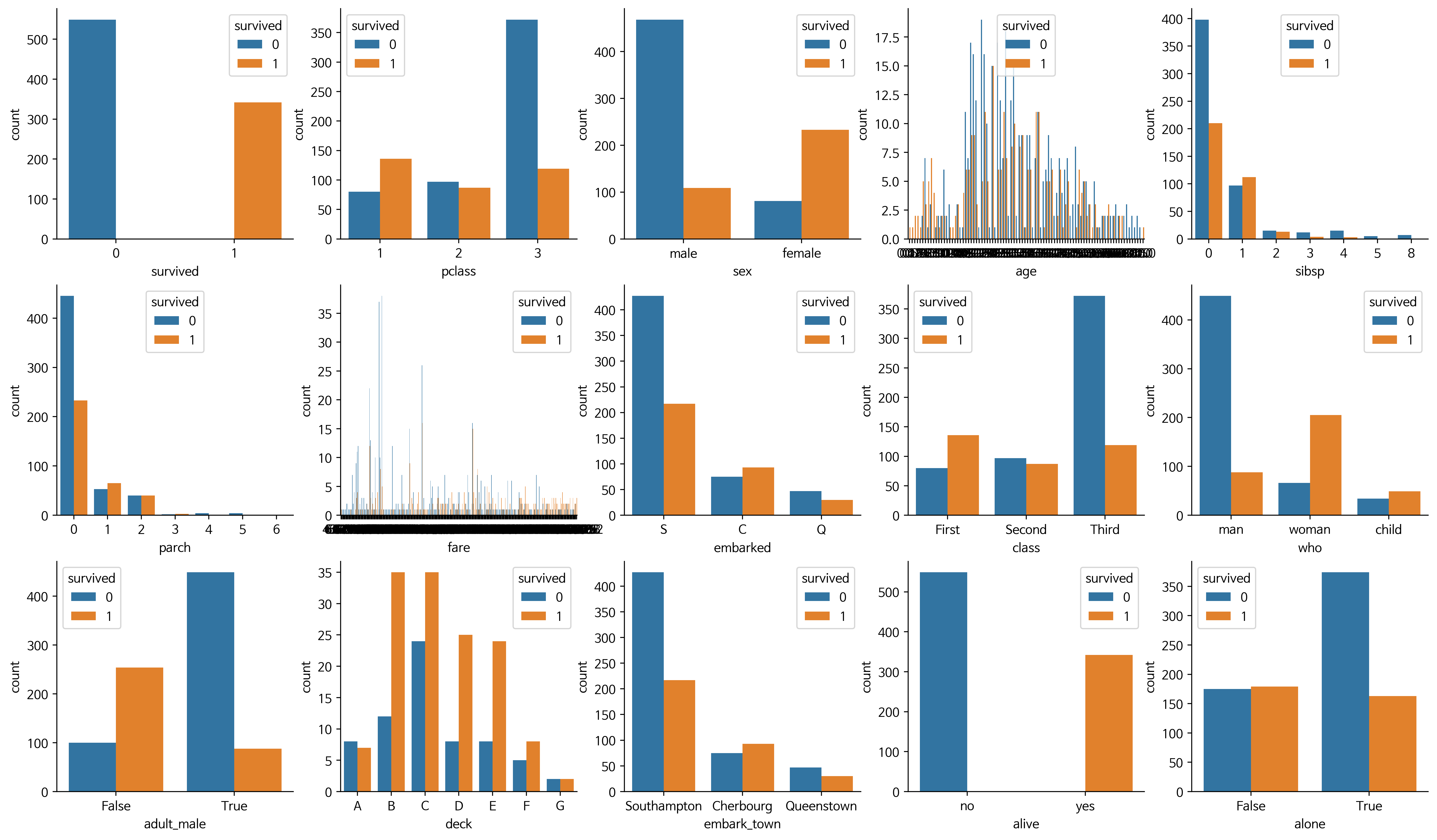
👀 그래프를 보고 넣고 뺄 것 고르기
데이터 전처리
df.isna().sum()
# 1) NaN이 많은 컬럼 및 중복 컬럼 삭제 -> deck(NaN 다수), embark_town(중복)
rdf = df.drop(['deck','embark_town'], axis=1)
rdf.info()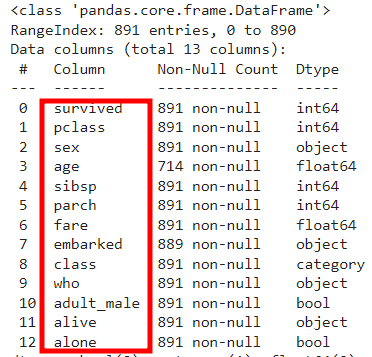
👀 지워짐
# 2) age 컬럼에 데이터가 없는 row(행) 삭제 -> age가 NaN인 177건만 삭제되도록
rdf = rdf.dropna(subset=['age'], how='any', axis=0)
rdf.info()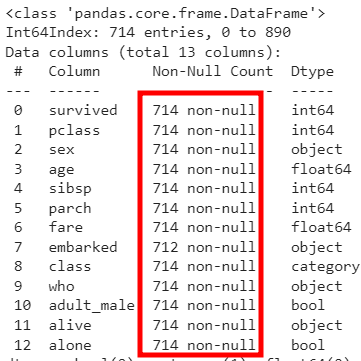
👀 삭제되어 줄어들었다
# embarked 승선도시 NaN 데이터 2건 어떻게채우지 ? -> 승선도시 중 가장 많이 출현한 도시로 지정
most_freq = rdf['embarked'].value_counts(dropna=True).idxmax()
most_freq # -> 'S'
# 'S'이므로 NaN 데이터 'S'로 지정
# embarked 열의 NaN값을 승선도시 중 가장 많이 출현한 도시로 채우기
rdf['embarked'].fillna(most_freq, inplace=True)
rdf.isna().sum() 👀 NaN값이 없음을 볼 수 있음!
👀 NaN값이 없음을 볼 수 있음!
# 4) 학습에 필요한 컬럼을 선택
# 생존여부, 객실 등급, 성별, 나이, 형제/자매수, 부모/자녀수, 탑승도시
ndf = rdf[['survived','pclass','sex','age','sibsp','parch','embarked']]
ndf.head()
# 5) 문자로 되어있는 값 -변환 > 인코딩 -> 원핫인코딩(범주형 데이터를 머신러닝 모델이 인식할 수 있도록 숫자형으로 변환)
# ex) male [1,0], female [0,1]
# ex) S[1,0,0],C[0,1,0],Q[0,0,1]
# 5-1) onehot 인코딩
onehot_sex = pd.get_dummies(ndf['sex'])
onehot_embarked = pd.get_dummies(ndf['embarked'])
# 5-2) ndf 데이터프레임에 연결
ndf = pd.concat([ndf, onehot_sex],axis=1)
ndf = pd.concat([ndf, onehot_embarked],axis=1)
# 6) 기존 컬럼삭제
ndf.drop(['sex','embarked'], axis=1, inplace=True)
ndf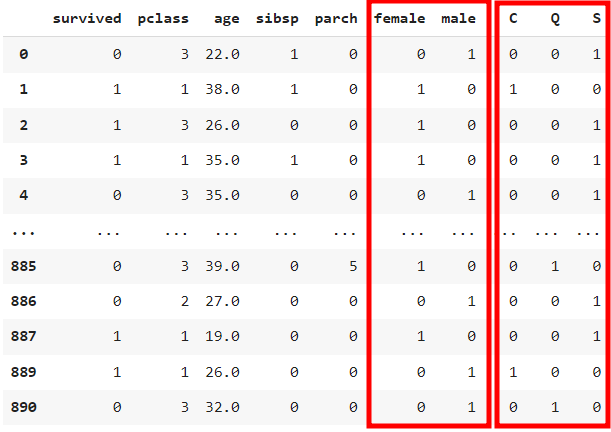
데이터 분리하기
X = ndf[['pclass', 'age', 'sibsp', 'parch', 'female', 'male', 'C', 'Q', 'S']]
y = ndf['survived']
# X (feature, 독립변수) 값을 정규화 -> 0~1 사이로 값을 줄여주는 작업 -> 스케일링(범위조정)
from sklearn import preprocessing
X = preprocessing.StandardScaler().fit(X).transform(X)
X
# train, test set으로 분리(7:3)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y , test_size=0.3, random_state=7)
print('trains shape : ', X_train.shape)
print('test shape : ', X_test.shape)SVM 분류모델
from sklearn import svm
# 모델 객체 생성 kernel = 'rbf'
# 벡터 공간을 맵핑하는함수 -> 선형(linear), 다항식(poly), 가우시안 RBF(rbf), 시그모이드(sigmoid)
svm_model = svm.SVC(kernel = 'rbf')
# 모델 학습
svm_model.fit(X_train, y_train)
print('훈련 세트 점수 : {:.8f}'.format(svm_model.score(X_train, y_train)))
print('테스트 세트 점수 : {:.8f}'.format(svm_model.score(X_test, y_test)))
# 모델 학습 결과
from sklearn import metrics
y_pred = svm_model.predict(X_test) # 문제풀어봐
print('accuracy : ', metrics.accuracy_score(y_test, y_pred))
print('precision : ', metrics.precision_score(y_test, y_pred))
print('recall : ', metrics.recall_score(y_test, y_pred))
print('f1s : ', metrics.f1_score(y_test, y_pred))accuracy : 0.8046511627906977
precision : 0.873015873015873
recall : 0.6179775280898876
f1s : 0.7236842105263157
- 선형 모델
주요 매개변수 - 회귀변수 -> alpha, LinearSVC, LogisticRegression -> C
alpha값이 클 수록, C값이 작을수록 모델이 단순해짐
보통 C와 alpha는 로그 스케일로 최적치를 정함 - L1, L2 규제 중 무엇을 사용할지 정해야함
중요한 특성이 많지 않음 -> L1
중요한 특성이 많음 -> L2
L1 규제 : 모델의 해석이 중요한 요소일 때도 사용할 수 있다.
몇가지 특성만 사용하므로 해당 모델에 중요한 특성이 무엇이고 효과가 어느 정도인지 설명하기 쉬움 - 선형 모델은 학습 속도가 빠름
- 회귀와 분류에서 본 공식을 사용해 예측이 어떻게 만들어지는지 비교적 쉽게 이해




