◾Principal Component Analysis
Principal Component Analysis(PCA): 데이터 집합 내에 존재하는 각 데이터의 차이를 가장 잘 나타내 주는 요소를 찾아내는 방법- 통계 데이터 분석(주성분 찾기), 데이터 압축(차원 감소), 노이즈 제거 등 다양한 분야에서 사용
- 차원 축소(dimensionality reduction)와 변수 추출(feature extraction) 기법으로 널리 쓰이고 있는 주성분 분석(Principal Component Analysis)
- 분산(Variance)을 최대한 보존하면서 서로 직교하는 새 기저(축)을 찾아, 고차원 공간의 표본들을 선형 연관성이 없는 저차원 공간으로 변환하는 기법
변수 추출은 기존 변수를 조합해 새로운 변수를 만드는 기법으로 변수 선택과 구분해야한다.
- PCA
- 데이터를 어떤 벡터에 정사영시켜 차원을 낮출 수 있다.
- 벡터를 이용해 데이터 다시 표현하기
- 행렬 : n X m => (PCA) => 새 행렬 : n X k
- 데이터를 변환시켰을 때, 어떤 벡터를 선정하면 본래 데이터 구조를 가장 잘 유지할 수 있을까
- 데이터를 새로운 축으로 표현하는 것

- 차원이 많은 경우 간단하게 표현해볼 수 있다.
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
sns.set_style('whitegrid')- 간단한 데이터 실습
rng = np.random.RandomState(13)
# (2, 2)행렬, (2, 200) 정규분포 행렬의 곱
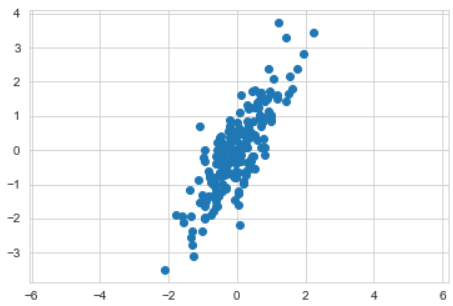
X = np.dot(rng.rand(2, 2), rng.randn(2, 200)).T
X.shapeplt.scatter(X[:, 0], X[:, 1])
plt.axis('equal')
plt.show()- PCA 학습
from sklearn.decomposition import PCA
# n_components : 표현할 주성분의 수
pca = PCA(n_components=2, random_state = 13)
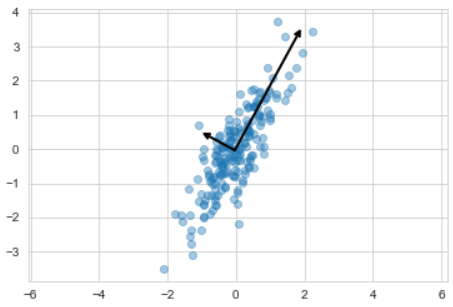
pca.fit(X)# 벡터 값
pca.components_# 분산값(설명력)
pca.explained_variance_# 평균값(일종의 좌표 원점값)
pca.mean_- 주성분 벡터 그리기
def draw_vector(v0, v1, ax=None):
ax = ax or plt.gca()
arrowprops = dict(arrowstyle='->',
linewidth=2, color='black',
shrinkA=0, shrinkB=0)
ax.annotate('', v1, v0, arrowprops=arrowprops)plt.scatter(X[:, 0], X[:,1], alpha=0.4)
for length, vector in zip(pca.explained_variance_, pca.components_):
v = vector * 3 * np.sqrt(length)
draw_vector(pca.mean_, pca.mean_ + v)
plt.axis('equal')
plt.show()- 주성분을 이용한 축 변경
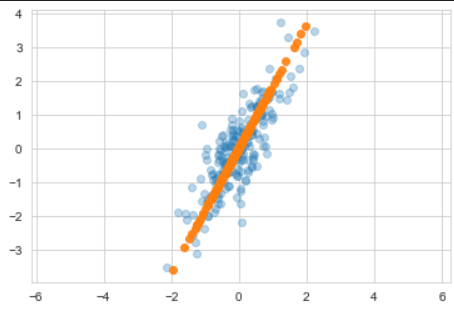
- 주성분을 찾은 다음 주축으로 변경할수도 있다.
pca = PCA(n_components=1, random_state = 13)
pca.fit(X)
X_pca = pca.transform(X)
print(pca.components_)
print(pca.explained_variance_)
print(pca.mean_)# 변환한 값을 원래 값에 매칭시킬 수 있다.
X_new = pca.inverse_transform(X_pca)
plt.scatter(X[:, 0], X[:, 1], alpha=0.3)
plt.scatter(X_new[:, 0], X_new[:, 1], alpha=0.9)
plt.axis('equal')
plt.show()◾PCA - Wine Data
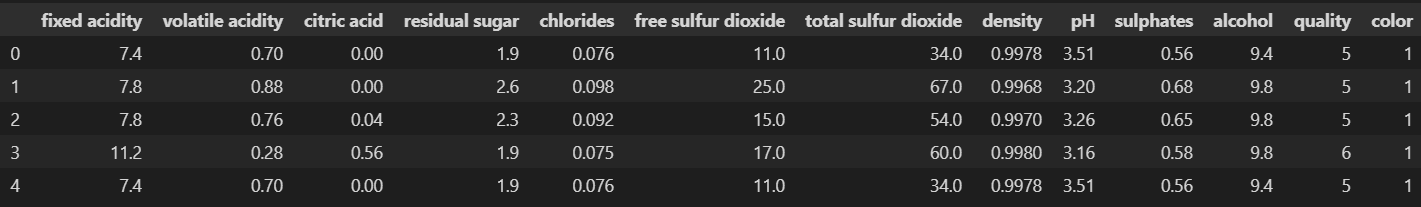
- 와인 데이터 읽기
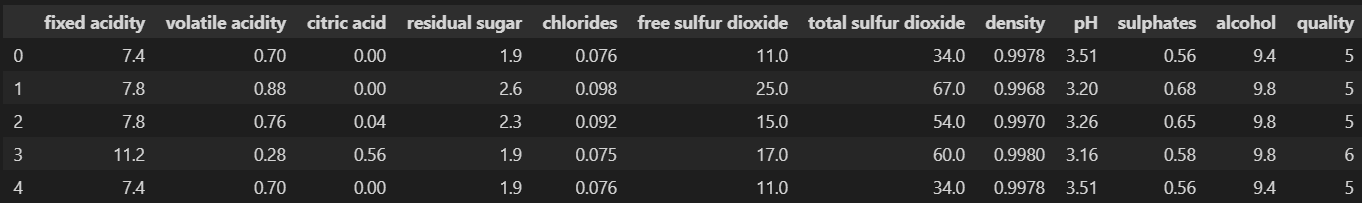
wine = pd.read_csv('../data/02/wine.csv', sep=',', index_col=0)
wine.head()- 와인 색상 분류(red/white)
wine_y = wine['color']
wine_X = wine.drop(['color'], axis=1)
wine_X.head()- StandardScaler
wine_ss = StandardScaler().fit_transform(wine_X)
wine_ss[:3]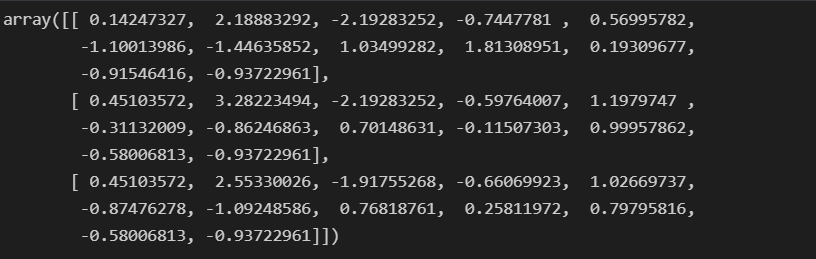
wine_ss.shape- PCA 적용(2개의 주성분)
def print_variance_ratio(pca):
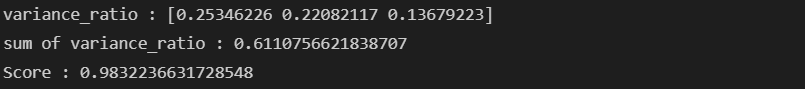
print('variance_ratio : {}'.format(pca.explained_variance_ratio_))
print('sum of variance_ratio : {}'.format(np.sum(pca.explained_variance_ratio_)))pca_wine, pca = get_pca_data(wine_ss, n_components=2)
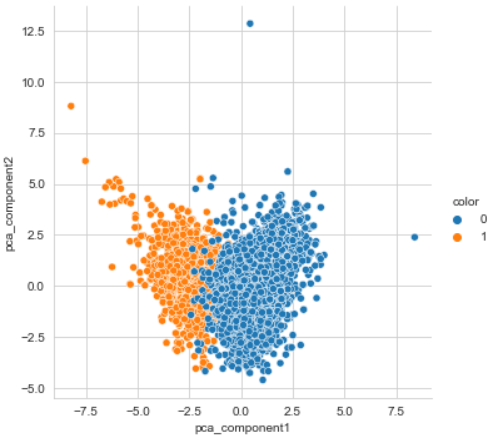
pca_wine.shape- 설명력이 작은 것을 볼 수 있다.
print_variance_ratio(pca)- 시각화
pca_columns = ['pca_component1', 'pca_component2']
pca_wine_pd = pd.DataFrame(pca_wine, columns=pca_columns)
pca_wine_pd['color'] = wine_y.values
sns.pairplot(pca_wine_pd, hue='color', height=5,
x_vars=['pca_component1'], y_vars=['pca_component2']);- RandomForest
- 비슷한 결과를 볼 수 있다.
# 원 데이터
rf_scores(wine_ss, wine_y)# 주성분 2개
pca_X = pca_wine_pd[['pca_component1', 'pca_component2']]
rf_scores(pca_X, wine_y)- PCA 적용(3개의 주성분)
pca_wine, pca = get_pca_data(wine_ss, n_components=3)
print_variance_ratio(pca)
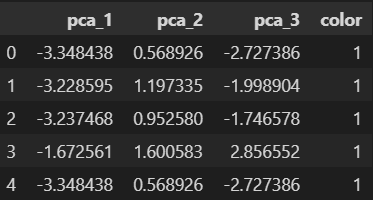
pca_cols = ['pca_1', 'pca_2', 'pca_3']
pca_wine_pd = pd.DataFrame(pca_wine, columns=pca_cols)
# 주성분 3개
pca_X = pca_wine_pd[pca_cols]
rf_scores(pca_X, wine_y)# 결과 정리
pca_wine_plot = pca_X
pca_wine_plot['color'] = wine_y.values
pca_wine_plot.head()3D로 그리기
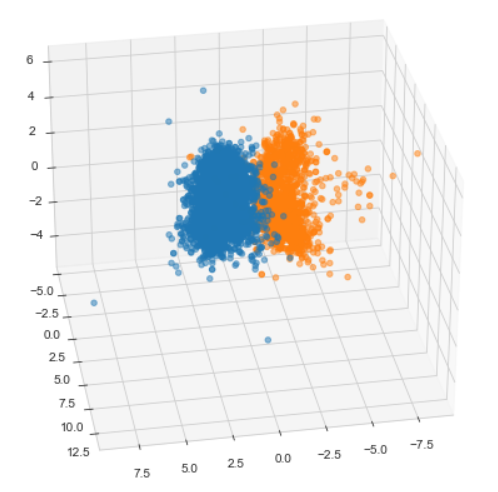
from mpl_toolkits.mplot3d import Axes3D
markers=['^', 'o']
fig = plt.figure(figsize=(10, 8))
ax = fig.add_subplot(111, projection='3d')
for i, marker in enumerate(markers):
x_axis_data = pca_wine_plot[pca_wine_plot['color']==i]['pca_1']
y_axis_data = pca_wine_plot[pca_wine_plot['color']==i]['pca_2']
z_axis_data = pca_wine_plot[pca_wine_plot['color']==i]['pca_3']
ax.scatter(x_axis_data, y_axis_data, z_axis_data,
s = 20, alpha=0.5)
ax.view_init(30, 80)
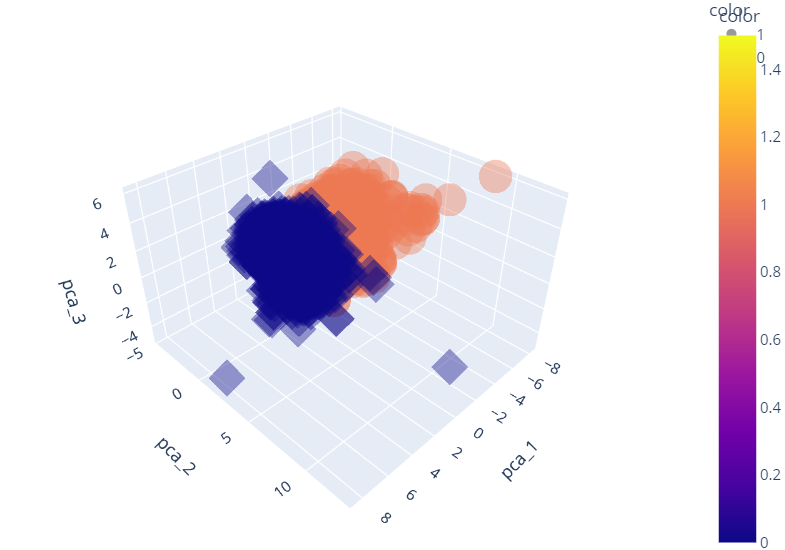
plt.show()import plotly_express as px
fig = px.scatter_3d(pca_wine_plot,
x = 'pca_1', y='pca_2', z='pca_3',
color='color', symbol='color',
opacity=0.4)
fig.update_layout(margin=dict(l=0, r=0, b=0, t=0))
fig.show()