작성자 : 이재빈
PyTorch Geometric 을 이용하여 Graph Neural Networks 를 구현하고 학습하는 내용을 공부합니다.
PyTorch Geometric
torch-geometric : GNN implementation module
- Docs : https://pytorch-geometric.readthedocs.io/en/latest/modules/nn.html
- git : https://github.com/rusty1s/pytorch_geometric
- torch_geometric.nn : GNN Layer
- torch_geometric.utils : Loss / Evaluation Metrics / Utility Function
- torch_geometric.transforms : Transform Data
- networkx : Visualize Graph Structure
torch_geometric.data.Data : Graph Attributes
data.x: node feature matrix , [num_nodes, num_node_features]data.edge_index: graph connectivity , [2, num_edges]data.edge_attr: edge attribute matrix , [num_edges, num_edge_features]data.y: Graph or node targets- graph level : [num_nodes, *]
- node label : [1, *]
Setup
# install
!pip install --verbose --no-cache-dir torch-scatter
!pip install --verbose --no-cache-dir torch-sparse
!pip install --verbose --no-cache-dir torch-cluster
!pip install torch-geometric
!pip install tensorboardX
!wget https://bin.equinox.io/c/4VmDzA7iaHb/ngrok-stable-linux-amd64.zip
!unzip ngrok-stable-linux-amd64.zipimport torch
import torch.nn as nn
import torch.nn.functional as F
import torch_geometric.nn as pyg_nn # GNN module
import torch_geometric.utils as pyg_utils # GNN Utility Function
import torch_geometric.transforms as T
import time
from datetime import datetime
import networkx as nx # visualize Graph Structure
import numpy as np
import torch
import torch.optim as optim
# dataset
from torch_geometric.datasets import TUDataset
from torch_geometric.datasets import Planetoid
from torch_geometric.data import DataLoader
# visualize
from tensorboardX import SummaryWriter
from sklearn.manifold import TSNE
import matplotlib.pyplot as pltDefine the Model
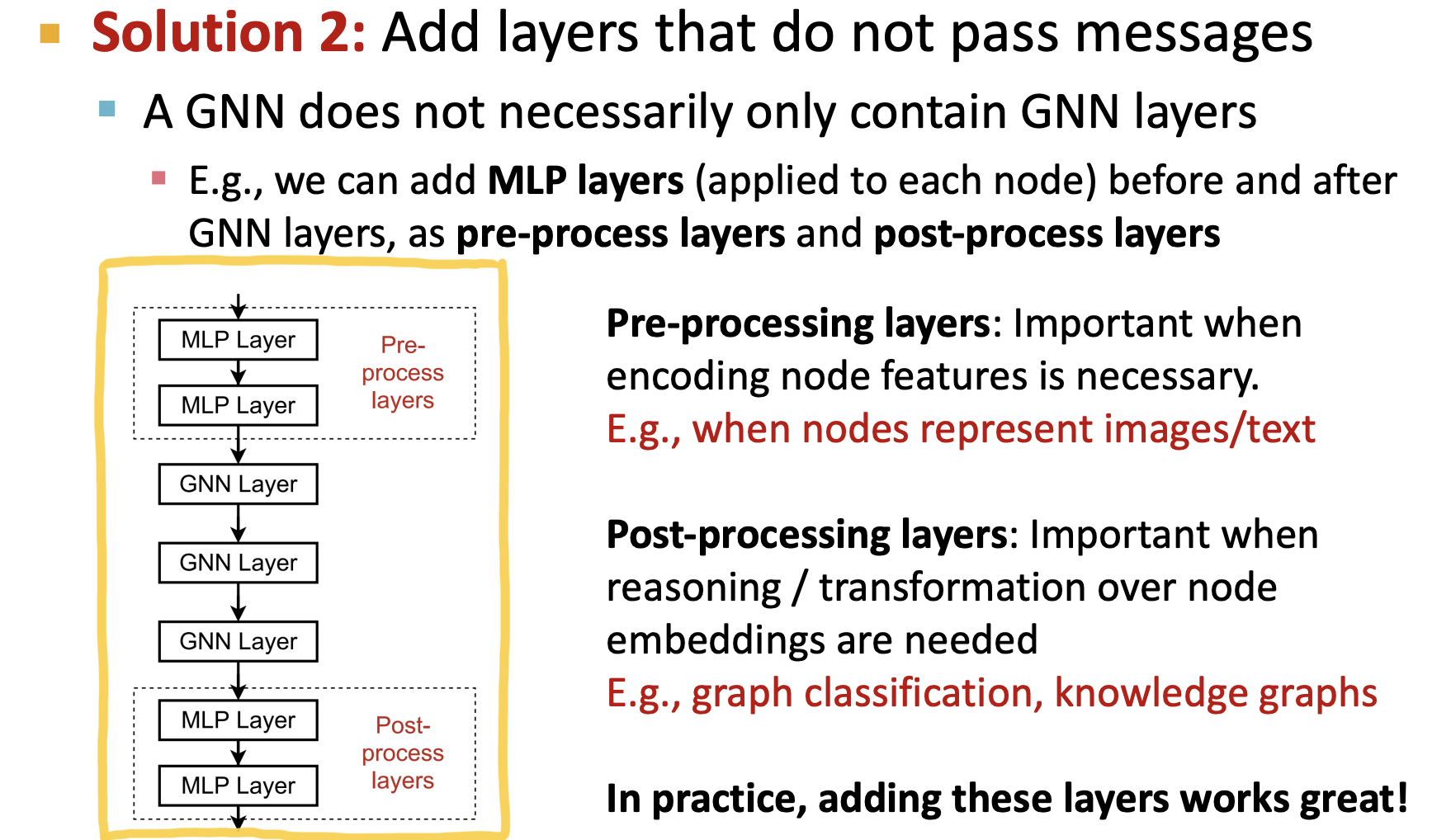
1. torch_geometric
class GNNStack(nn.Module): # stacking of Graph Convolutions
def __init__(self, input_dim, hidden_dim, output_dim, task='node'):
super(GNNStack, self).__init__()
self.task = task
# nn.ModuleList() : nn.Module()을 list로 정리! 각 layer를 list로 전달하고, layer의 iterator를 만듭니다.
self.convs = nn.ModuleList()
self.convs.append(self.build_conv_model(input_dim, hidden_dim))
self.lns = nn.ModuleList()
self.lns.append(nn.LayerNorm(hidden_dim))
self.lns.append(nn.LayerNorm(hidden_dim))
for l in range(2):
self.convs.append(self.build_conv_model(hidden_dim, hidden_dim))
# post-message-passing
self.post_mp = nn.Sequential(
nn.Linear(hidden_dim, hidden_dim), nn.Dropout(0.25),
nn.Linear(hidden_dim, output_dim))
if not (self.task == 'node' or self.task == 'graph'):
raise RuntimeError('Unknown task.')
self.dropout = 0.25
self.num_layers = 3
# task에 따른 convolution layer를 만들어 줍니다.
def build_conv_model(self, input_dim, hidden_dim):
# refer to pytorch geometric nn module for different implementation of GNNs.
if self.task == 'node': # node classification
return pyg_nn.GCNConv(input_dim, hidden_dim)
# return CustomConv(input_dim, hidden_dim) : run my method
else:
return pyg_nn.GINConv(nn.Sequential(nn.Linear(input_dim, hidden_dim),
nn.ReLU(), nn.Linear(hidden_dim, hidden_dim)))
def forward(self, data):
'''
x : feature matrix (# of nodes, # of node feature dim)
edge_index : sparse adj list, 연결된 edge에 대한 node 저장
ex. node 1 : [1,4,6]
batch : (array) batch마다 node 개수가 달라지므로 -> 어떤 node가 어떤 graph에 속하는지에 대한 정보 저장
ex. [1,1,1,1,1] : 5 nodes in graph 1 , [2,2,2] : 3 nodes in graph 2
'''
x, edge_index, batch = data.x, data.edge_index, data.batch
if data.num_node_features == 0: # feature 없으면 -> constant
x = torch.ones(data.num_nodes, 1)
# Neural Network
for i in range(self.num_layers):
x = self.convs[i](x, edge_index) # Conv Layer
emb = x
x = F.relu(x)
x = F.dropout(x, p=self.dropout, training=self.training)
if not i == self.num_layers - 1:
x = self.lns[i](x)
if self.task == 'graph': # mean pooling : average all the nodes
x = pyg_nn.global_mean_pool(x, batch)
x = self.post_mp(x)
return emb, F.log_softmax(x, dim=1)
def loss(self, pred, label):
return F.nll_loss(pred, label) # negative log-likelihood 2. Custom Model
class CustomConv(pyg_nn.MessagePassing): # inherenting from MessagePassing
def __init__(self, in_channels, out_channels):
super(CustomConv, self).__init__(aggr='add') # Neighborhood Aggregation : Mean, Max, Add, ...
self.lin = nn.Linear(in_channels, out_channels)
self.lin_self = nn.Linear(in_channels, out_channels)
def forward(self, x, edge_index):
'''
x : feature matrix
edge_index : connectivity, Adj list in the edge index
'''
# x has shape [N, in_channels]
# edge_index has shape [2, E]
# original code
# Add self-loops to the adjacency matrix : neighbor + self
# pyg_utils.add_self_loops(edge_index, num_nodes = x.size(0)) # A + I
# 여기에서는 remove self-loops : skip layer on top of that
edge_index, _ = pyg_utils.remove_self_loops(edge_index)
# Transform node feature matrix.
self_x = self.lin_self(x) # B
# x = self.lin(x) # W
return self_x + self.propagate(edge_index, size=(x.size(0), x.size(0)), x=self.lin(x))
def message(self, x_i, x_j, edge_index, size):
'''
GCN : D^(-1/2)*A*D(1/2)*W*X
x_i : self
x_j : neighborhood
'''
# Compute messages
# x_j has shape [E, out_channels]
row, col = edge_index
deg = pyg_utils.degree(row, size[0], dtype=x_j.dtype)
deg_inv_sqrt = deg.pow(-0.5)
norm = deg_inv_sqrt[row] * deg_inv_sqrt[col]
return x_j
def update(self, aggr_out):
# aggr_out has shape [N, out_channels] : add additional layer after message passing
# GraphSAGE : L2 Normalization
# F.normalize(aggr_out, p=2, dim=-1)
return aggr_out# Custom Model 을 사용하는 경우, GNNStack class 내의 build_conv_model return 값을 다음과 같이 수정해 주면 됩니다.
def build_conv_model(self, input_dim, hidden_dim):
# refer to pytorch geometric nn module for different implementation of GNNs.
if self.task == 'node':
# return pyg_nn.GCNConv(input_dim, hidden_dim)
return CustomConv(input_dim, hidden_dim) # run my method
else:
return pyg_nn.GINConv(nn.Sequential(nn.Linear(input_dim, hidden_dim),
nn.ReLU(), nn.Linear(hidden_dim, hidden_dim)))Model

Training Setup
Train
def train(dataset, task, writer):
if task == 'graph':
data_size = len(dataset)
loader = DataLoader(dataset[:int(data_size * 0.8)], batch_size=64, shuffle=True)
test_loader = DataLoader(dataset[int(data_size * 0.8):], batch_size=64, shuffle=True)
else:
test_loader = loader = DataLoader(dataset, batch_size=64, shuffle=True)
# build model
model = GNNStack(max(dataset.num_node_features, 1), 32, dataset.num_classes, task=task)
opt = optim.Adam(model.parameters(), lr=0.01)
# train
for epoch in range(200):
total_loss = 0
model.train()
for batch in loader:
#print(batch.train_mask, '----')
opt.zero_grad()
embedding, pred = model(batch)
label = batch.y
if task == 'node':
pred = pred[batch.train_mask]
label = label[batch.train_mask]
loss = model.loss(pred, label)
loss.backward()
opt.step()
total_loss += loss.item() * batch.num_graphs
total_loss /= len(loader.dataset)
writer.add_scalar("loss", total_loss, epoch)
if epoch % 10 == 0:
test_acc = test(test_loader, model)
print("Epoch {}. Loss: {:.4f}. Test accuracy: {:.4f}".format(
epoch, total_loss, test_acc))
writer.add_scalar("test accuracy", test_acc, epoch)
return modelValidation / Test
def test(loader, model, is_validation=False):
model.eval()
correct = 0
for data in loader:
with torch.no_grad():
emb, pred = model(data)
pred = pred.argmax(dim=1)
label = data.y
# mask 를 통해 validation, test 결정
if model.task == 'node':
mask = data.val_mask if is_validation else data.test_mask
# node classification: only evaluate on nodes in test set
pred = pred[mask]
label = data.y[mask]
correct += pred.eq(label).sum().item()
if model.task == 'graph':
total = len(loader.dataset)
else:
total = 0
for data in loader.dataset:
total += torch.sum(data.test_mask).item()
return correct / totalTraining the Model
# Setting TensorboardX in Colab
get_ipython().system_raw(
'tensorboard --logdir {} --host 0.0.0.0 --port 6006 &'
.format("./log")
)
get_ipython().system_raw('./ngrok http 6006 &')
!curl -s http://localhost:4040/api/tunnels | python3 -c \
"import sys, json; print(json.load(sys.stdin)['tunnels'][0]['public_url'])"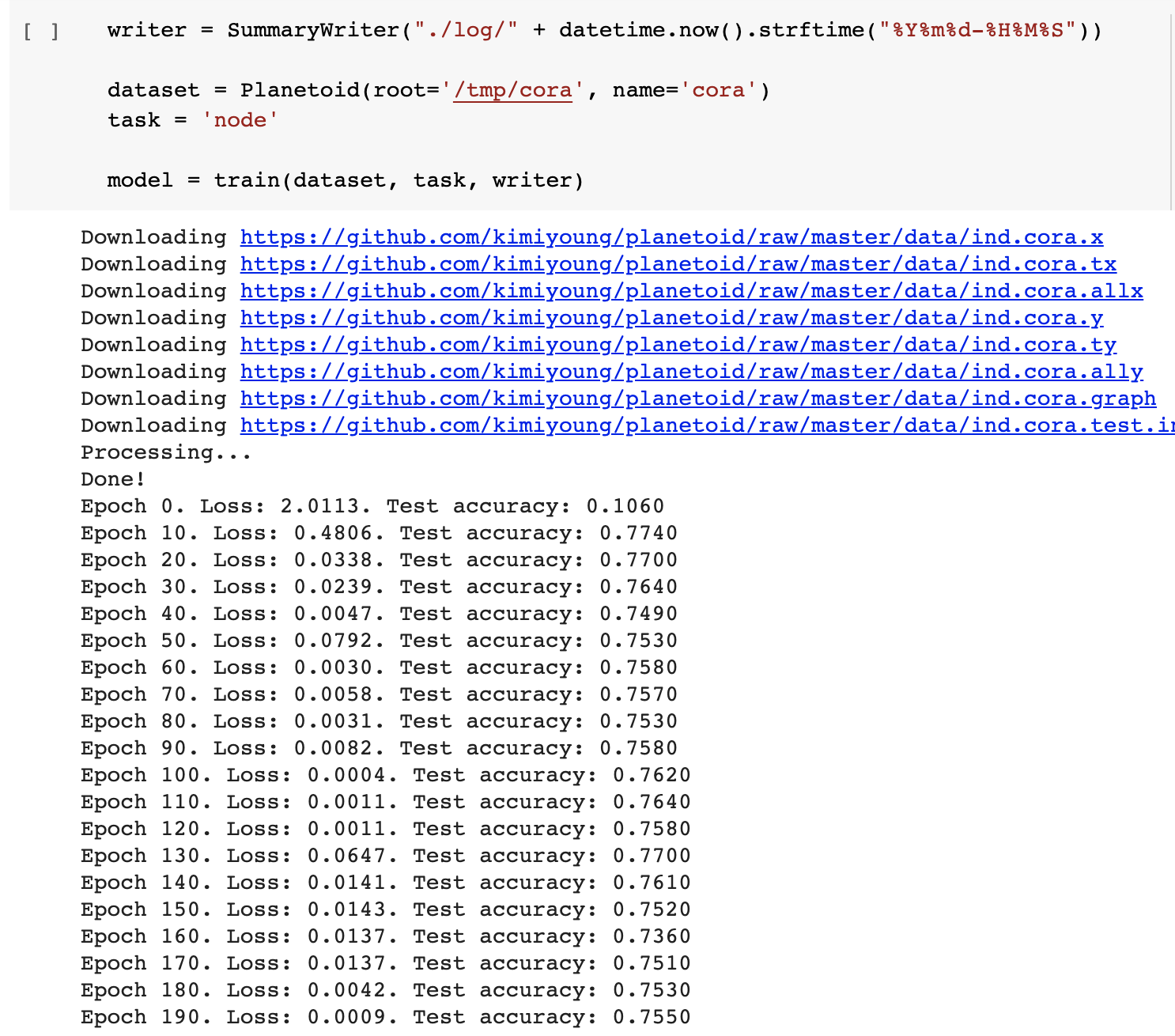
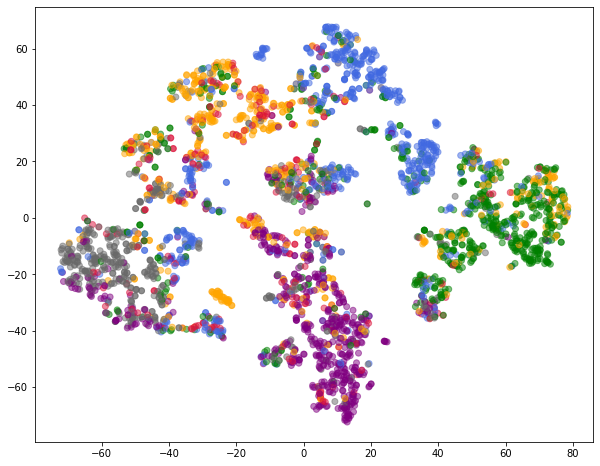
Visualize Node Embeddings
color_list = ["crimson", "orange", "green", "royalblue", "purple", "dimgrey", "gold"]
loader = DataLoader(dataset, batch_size=64, shuffle=True)
embs = []
colors = []
for batch in loader:
emb, pred = model(batch)
embs.append(emb)
colors += [color_list[y] for y in batch.y]
embs = torch.cat(embs, dim=0)
xs, ys = zip(*TSNE().fit_transform(embs.detach().numpy()))
plt.figure(figsize=(10, 8))
plt.scatter(xs, ys, color=colors, alpha=0.5)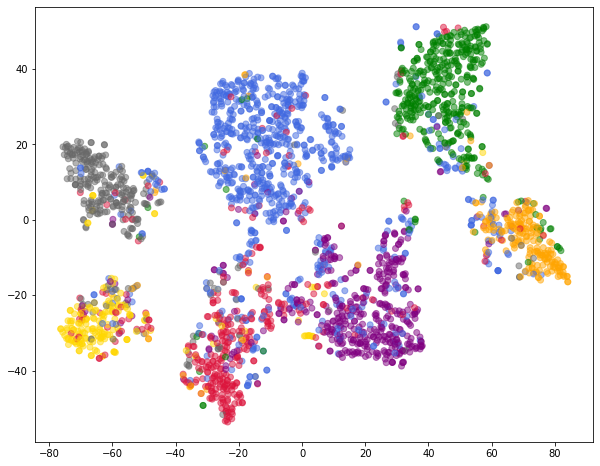
Learning Unsupervised Embeddings with Graph AutoEncoders
# VGAE : variational graph auto-encoder
# Knowledge Graph, Graph Reasoning
class Encoder(torch.nn.Module):
'''
Encoder : Graph Conv to get embeddings
Decoder : inner product -> 2개 node 사이의 값이 크면, there's a likely link between them
'''
def __init__(self, in_channels, out_channels):
super(Encoder, self).__init__()
self.conv1 = pyg_nn.GCNConv(in_channels, 2 * out_channels, cached=True)
self.conv2 = pyg_nn.GCNConv(2 * out_channels, out_channels, cached=True)
def forward(self, x, edge_index):
x = F.relu(self.conv1(x, edge_index))
return self.conv2(x, edge_index)
def train(epoch):
model.train()
optimizer.zero_grad()
z = model.encode(x, train_pos_edge_index)
loss = model.recon_loss(z, train_pos_edge_index) # reconstruction loss
loss.backward()
optimizer.step()
writer.add_scalar("loss", loss.item(), epoch)
def test(pos_edge_index, neg_edge_index):
model.eval()
with torch.no_grad():
z = model.encode(x, train_pos_edge_index)
return model.test(z, pos_edge_index, neg_edge_index)writer = SummaryWriter("./log/" + datetime.now().strftime("%Y%m%d-%H%M%S"))
dataset = Planetoid("/tmp/citeseer", "Citeseer", transform = T.NormalizeFeatures())
data = dataset[0]
channels = 16
dev = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print('CUDA availability:', torch.cuda.is_available())# encoder: written by us; decoder: default (inner product)
model = pyg_nn.GAE(Encoder(dataset.num_features, channels)).to(dev)
labels = data.y
data.train_mask = data.val_mask = data.test_mask = data.y = None
# data = model.split_edges(data) # split_edges 안 돌아가서 변경!
data = pyg_utils.train_test_split_edges(data) # construct positive/negative edges (for negative sampling!)
x, train_pos_edge_index = data.x.to(dev), data.train_pos_edge_index.to(dev)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
for epoch in range(1, 201):
train(epoch)
auc, ap = test(data.test_pos_edge_index, data.test_neg_edge_index)
writer.add_scalar("AUC", auc, epoch)
writer.add_scalar("AP", ap, epoch)
if epoch % 10 == 0:
print('Epoch: {:03d}, AUC: {:.4f}, AP: {:.4f}'.format(epoch, auc, ap))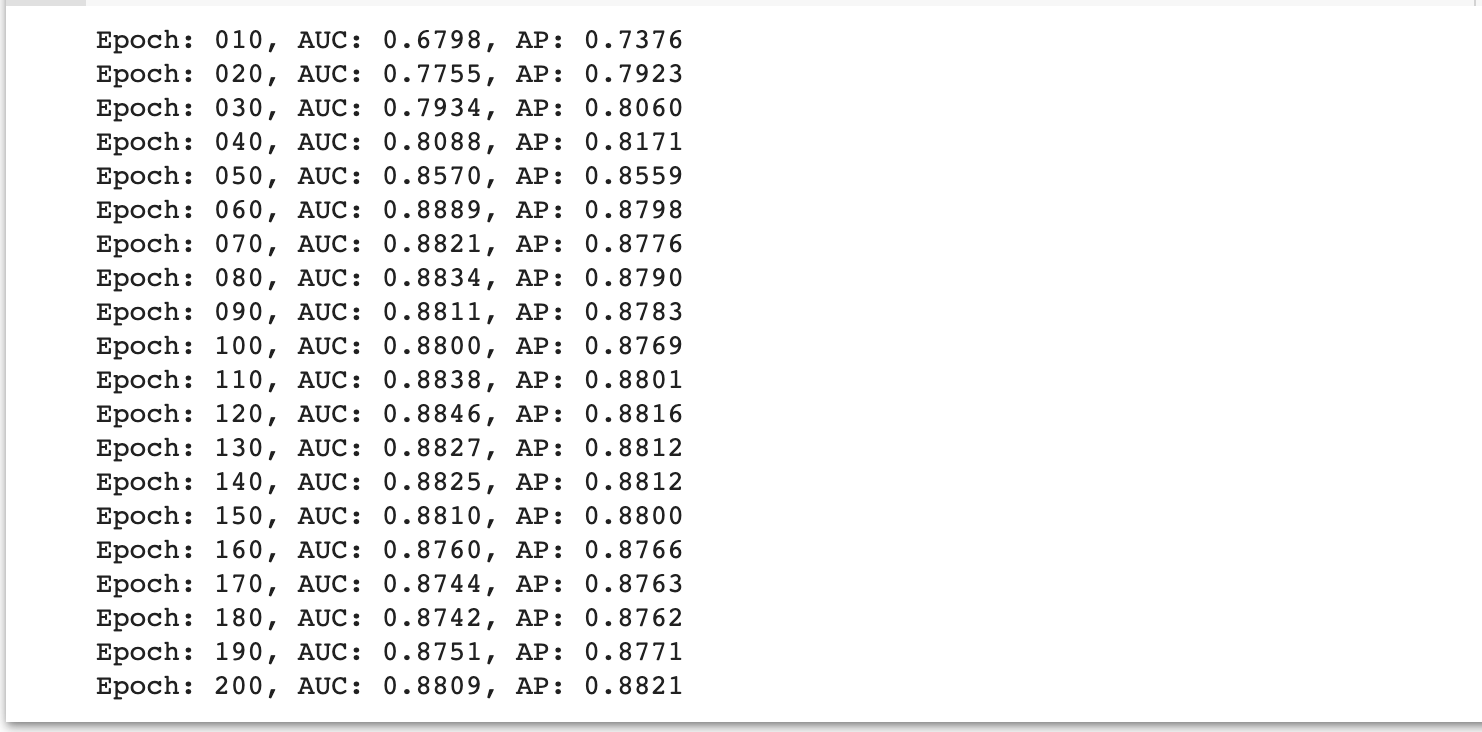
model.eval()
z = model.encode(x, train_pos_edge_index)
colors = [color_list[y] for y in labels]
xs, ys = zip(*TSNE().fit_transform(z.cpu().detach().numpy()))
plt.figure(figsize=(10, 8))
plt.scatter(xs, ys, color=colors, alpha=0.5)
plt.show()