
평가 📈
머신러닝 모델은 여러가지 방법으로 예측 성능을 평가할 수 있음. 성능 평가 지표는 일반적으로 모델이 분류나 회귀냐에 따라 여러 종류로 나뉨.
분류의 성능 평가 지표
🔹 정확도
🔹 오차행렬
🔹 정밀도
🔹 재현율
🔹 F1 스코어
🔹 ROC AUC
01. 정확도
정확도는 실제 데이터에서 예측 데이터가 얼마나 같은지를 판단하는 지표

- 단순한 Classifier 생성
from sklearn.base import BaseEstimator
class MyDummyClassifier(BaseEstimator):
# fit() 메서드는 아무것도 학습하지 않음.
def fit(self, X, y=None):
pass
# predict() 메서드는 단순히 Sex 피처가 1이면 0, 그렇지 않으면 1로 예측함.
def predict(self, X):
pred = np.zeros((X.shape[0], 1))
for i in range(X.shape[0]):
if X['Sex'].iloc[i] == 1:
pred[i] = 0
else:
pred[i] = 1
return pred타이타닉 생존자 예측
## 생성된 MyDummyClassifier를 이용해 타이타닉 생존자 예측 수행
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import LabelEncoder
## Null 처리 함수
def fillna(df):
df['Age'].fillna(df['Age'].mean(), inplace=True)
df['Cabin'].fillna('N', inplace=True)
df['Embarked'].fillna('N', inplace=True)
df['Fare'].fillna(0, inplace=True)
return df
## 머신러닝에 불필요한 피처 제거
def drop_features(df):
df.drop(['PassengerId', 'Name', 'Ticket'], axis=1, inplace=True)
return df
## Label Encoding 수행
def format_features(df):
df['Cabin'] = df['Cabin'].str[:1]
features = ['Cabin', 'Sex', 'Embarked']
for feature in features:
le = LabelEncoder()
le.fit(df[feature])
df[feature] = le.transform(df[feature])
return df
## 앞에서 실행한 Data Preprocessing 함수 호출
def transform_features(df):
df = fillna(df)
df = drop_features(df)
df = format_features(df)
return df
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
# 원본 데이터를 재로딩, 데이터 가공, 학습 데이터/테스트 데이터 분할
titanic_df = pd.read_csv('titanic_train.csv')
y_titanic_df = titanic_df['Survived']
X_titanic_df = titanic_df.drop('Survived', axis = 1)
X_titanic_df = transform_features(X_titanic_df)
#Survived 칼럼으로 target 설정
X_train, X_test, y_train, y_test = train_test_split(X_titanic_df, y_titanic_df, test_size=0.2, random_state=0)
# 위에서 생성한 Dummy Classifier를 이용해 학습/예측/평가 수행
myclf = MyDummyClassifier()
myclf.fit(X_train, y_train)
mypredictions = myclf.predict(X_test)
print('Dummy Classifier의 정확도는: {0:.4f}'.format(accuracy_score(y_test, mypredictions)))[Output]
Dummy Classifier의 정확도는: 0.7877이렇게 단순한 알고리즘은 정확도가 높으면 안됨
또한 데이터 분포도가 균일하지 않은 경우 높은 수치가 나타날 수 있는 것이 정확도 평가 지표의 맹점
02. 오차 행렬
오차 행렬에서 TN, FP, FN, TP 값을 다양하게 결합해 분류 모델 예측 성능의 오류가 어떠한 모습으로 발생하는지 알 수 있는 것
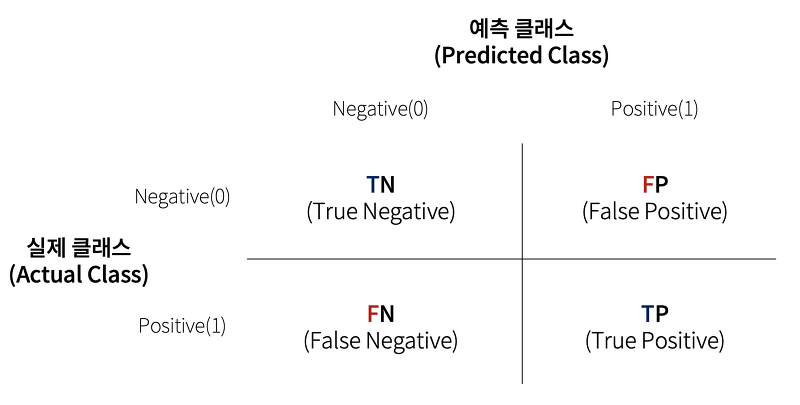
True/False 예측값과 실제값이 '같은가/틀린가'를 의미
Negative/Positive 예측 결과 값이 부정(0)/긍정(1)을 의미
- TN (True Negative)
예측 클래스 값과 실제 클래스 값이 Negative로 같음- FP (False Positive)
예측값은 Positive, 실제 값은 Negative로 다름- FN (False Negative)
예측값은 Negative, 실제 값은 Positive- TP (True Positive)
예측 클래스 값과 실제 클래스 값이 Positive로 같음
- 정확도
정확도는 예측값과 실제 값이 얼마나 동일한가에 대한 비율만으로 결정

중점적으로 찾아야 하는 매우 적은 수의 결괏값에 Positive를 설정, 그렇지 않은 경우 Negative를 설정 Positive 데이터 건수가 작기 때문에 Negative로 예측 정확도가 높아지는 경향이 발생 → 비대칭한 데이터 세트에서 Positive에 대한 예측 정확도를 판단하지 못한 채 Negative에 대한 예측 정확도만으로도 분류의 정확도가 매우 높게 나타남
03. 정밀도와 재현율
불균형 데이터 세트에서 선호됨(오차행렬의 한계 극복)
Positive 데이터 세트의 예측 성능에 좀 더 초점을 맞춘 평가 지표

- 정밀도
예측을 Positive로 한 대상 중에 예측과 실제 값이 Positive로 일치한 데이터의 비율
실제 Negative 음성인 데이터 예측을 Positive 양성으로 잘못 판단하게 되면 업무상 큰 영향이 발생하는 경우(아예 오류 차단하면 안되는 경우)
ex) 스팸메일 여부 판단 모델
- 재현율
실제 값이 Positive 대상 중에 예측과 실제 값이 Positive로 일치한 데이터의 비율
재현율이 중요 지표인 경우는 실제 Positive 양성 데이터를 Negative로 잘못 판단하게 되면 업무상 큰 영향이 발생하는 경우 (오류가 발생하면 안되는 경우)
ex) 암 판단 모델
get_clf_eval()
confusion_matrix, accuracy, precision, recall 평가를 한꺼번에 호출
from sklearn.metrics import accuracy_score, precision_score, recall_score, confusion_matrix
def get_clf_eval(y_test, pred):
confusion = confusion_matrix(y_test, pred)
accuracy = accuracy_score(y_test, pred)
precision = precision_score(y_test, pred)
recall = recall_score(y_test, pred)
print('오차 행렬')
print(confusion)
print('정확도: {0:.4f}, 정밀도: {1:.4f}, 재현율: {2:.4f}'.format(accuracy, precision, recall))- LogisticRegression으로 평가
이진 분류(solver = liblinear)로 시행
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
# 원본 데이터를 재로딩, 데이터 가공, 학습 데이터/테스트 데이터 분할
titanic_df = pd.read_csv('titanic_train.csv')
y_titanic_df = titanic_df['Survived']
X_titanic_df = titanic_df.drop('Survived', axis=1)
X_titanic_df = transform_features(X_titanic_df)
X_train, X_test, y_train, y_test = train_test_split(X_titanic_df, y_titanic_df, test_size = 0.20, random_state=11)
lr_clf = LogisticRegression(solver='liblinear')
lr_clf.fit(X_train, y_train)
pred = lr_clf.predict(X_test)
get_clf_eval(y_test, pred)[Output]
오차 행렬
[[108 10]
[ 14 47]]
정확도" 0.8659, 정밀도: 0.8246, 재현율: 0.7705- 정밀도/재현율 트레이드오프
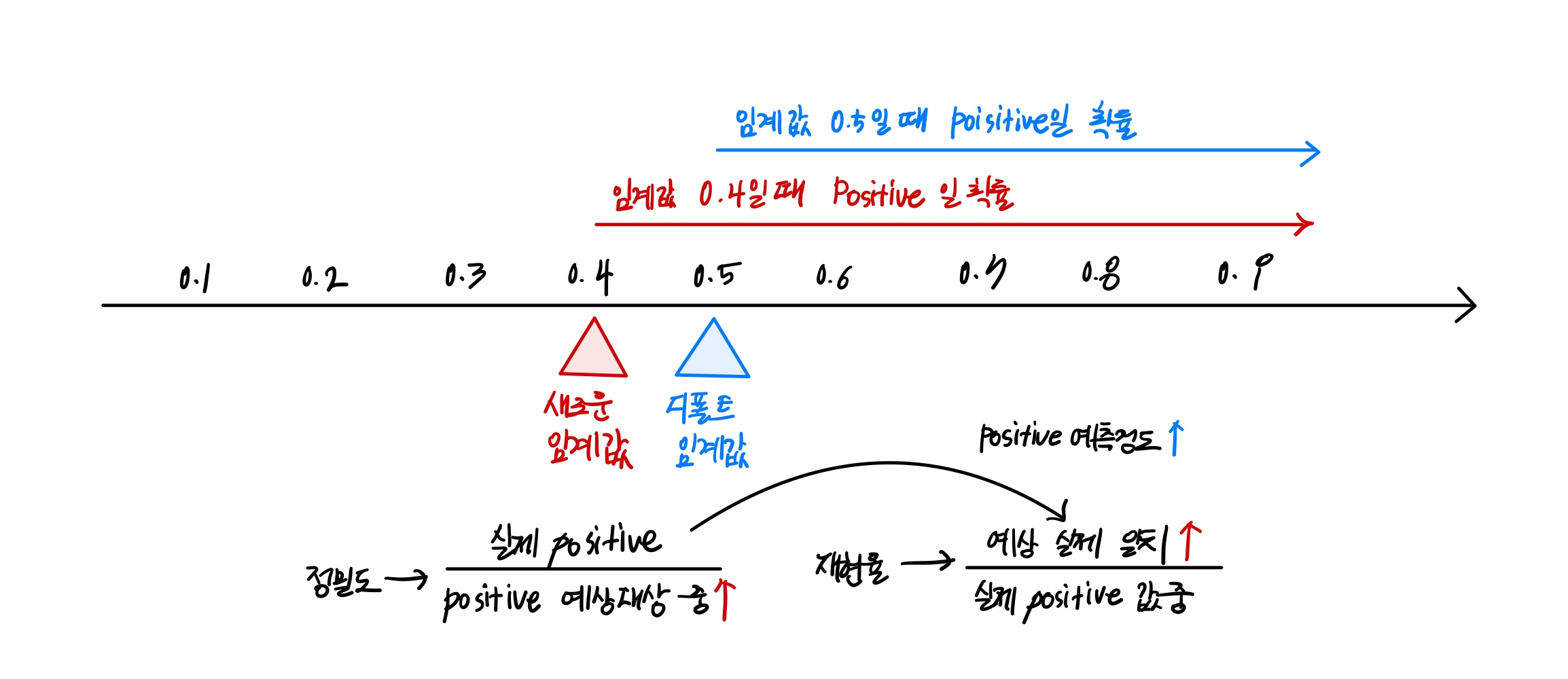
Threshold를 조절해 정밀도/재현율 조정 가능
정밀도와 재현율은 상호보완적인 평가 지표이기 때문에 어느 한쪽을 강제로 높이면 다른 하나의 수치는 떨어지기 때문에 트레이드 오프라 부름
일반적으로 이진분류에서는 공평하게 threshold를 공평하게 50%로 설정
predict_proba()
predict()메서드와 유사하지만 반환 값이 클래스 값이 아닌 예측 확률 결과
- predict probality랑 predict class를 함께 출력
pred_proba = lr_clf.predict_proba(X_test)
pred = lr_clf.predict(X_test)
print('pred_proba()결과 Shape : {0}'.format(pred_proba.shape))
print('pred_proba array에서 앞 3개만 샘플로 추출 \n:',pred_proba[:3])
# 예측 확률 array와 예측 결괏값 array를 병합(concatenate)해 예측 확률과 결괏값을 한눈에 확인
pred_proba_result = np.concatenate([pred_proba, pred.reshape(-1, 1)], axis=1)
print('두 개의 class 중에서 더 큰 확률을 클래스 값으로 예측 \n', pred_proba_result[:3])[Output]
pred_proba()결과 Shape : (179, 2)
pred_proba array에서 앞 3개만 샘플로 추출
: [[0.44935225 0.55064775]
[0.86335511 0.13664489]
[0.86429643 0.13570357]]
두 개의 class 중에서 더 큰 확률을 클래스 값으로 예측
[[0.44935225 0.55064775 1. ]
[0.86335511 0.13664489 0. ]
[0.86429643 0.13570357 0. ]]2개의 칼럼 중 더 큰 확률 값으로 predict() 메서드가 최종 예측
- Threshold 변화
from sklearn.preprocessing import Binarizer
# Binarizer의 threshold의 설정값. 분류 결정 임계값임.
custom_threshold = 0.5
# predict_proba() 반환값의 두 번째 칼럼, 즉 positive 클래스 칼럼 하나만 추출해 Binarizer를 적용
pred_proba_1 = pred_proba[:,1].reshape(-1, 1)
binarizer = Binarizer(threshold = custom_threshold).fit(pred_proba_1)
custom_predict = binarizer.transform(pred_proba_1)
get_clf_eval(y_test, custom_predict)
# Binarizer의 threshold의 설정값을 0.4로 설정. 즉 분류 결정 임계값을 0.5에서 0.4로 낮춤
custom_threshold = 0.4
pred_proba_1 = pred_proba[:,1].reshape(-1, 1)
binarizer = Binarizer(threshold=custom_threshold).fit(pred_proba_1)
custom_predict = binarizer.transform(pred_proba_1) #pred_proba_1은 제시하는 예측 결과
get_clf_eval(y_test, custom_predict)[Output]
오차 행렬
[[97 21]
[11 50]]
정확도: 0.8212, 정밀도: 0.7042, 재현율: 0.8197Threshold를 낮추니 재현율 값이 올라가고 정밀도가 떨어짐
Threshold은 Positive 예측값을 결정하는 확률의 기준이 됨. Threshold 값을 낮출수록 True 값이 많아짐
import matplotlib.pyplot as plt
import matplotlib.ticker as tiker
%matplotlib inline
def precision_recall_curve_plot(y_test, pred_proba_c1):
# threshold ndarray와 이 threshold에 따른 정밀도, 재현율 ndarray 추출.
precisions, recalls, thresholds = precision_recall_curve(y_test, pred_proba_c1)
# X축을 threshold값으로, Y축은 정밀도, 재현율 값으로 각각 Plot 수행. 정밀도는 점선으로 표시
plt.figure(figsize=(8, 6))
threshold_boundary = thresholds.shape[0]
plt.plot(thresholds, precisions[0:threshold_boundary], linestyle ='--', label='precision')
plt.plot(thresholds, recalls[0:threshold_boundary], label='recall')
# threshold 값 X 축의 Scale을 0.1 단위로 변경
start, end = plt.xlim()
plt.xticks(np.round(np.arange(start, end, 0.1), 2))
# x축. y축 label과 legend, 그리고 grid 설정
plt.xlabel('Threshold value')
plt.ylabel('Precision and Recall value')
plt.legend()
plt.grid()
plt.show()
precision_recall_curve_plot(y_test, lr_clf.predict_proba(X_test)[:,1])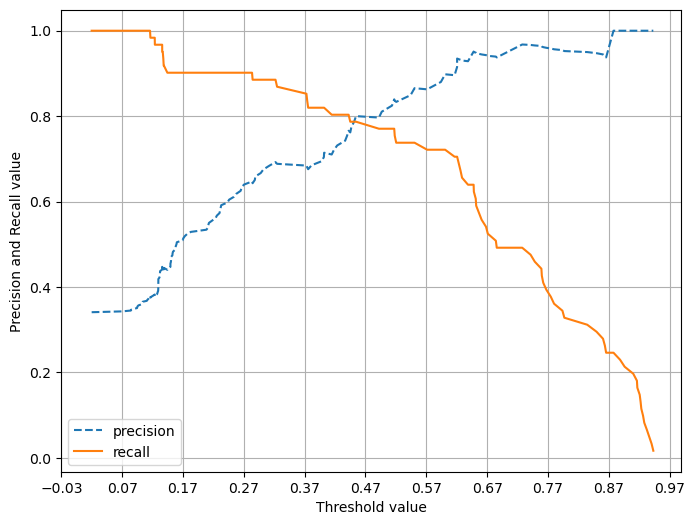
임계값이 낮을 수록 많은 수의 양성 예측으로 인해 재현율이 극도로 높
- 정밀도와 재현율의 맹점
두 개 수치를 상호보완 할 수 있는 수준에서 적용돼야 함. 단순히 하나의 성능 지표 수치를 높이기 위한 수단으로 사용돼서는 안 됨.
04. F1 스코어
F1 스코어는 정밀도와 재현율을 결합한 지표
정밀도 또는 정확도 쪽으로 치우치지 않는 수치를 나타낼 때 상대적으로 높은 값을 가짐
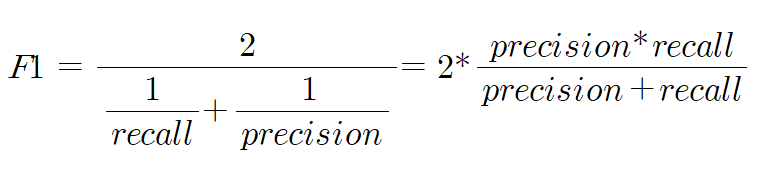
from sklearn.metrics import f1_score
f1 = f1_score(y_test, pred)
print('F1 스코어: {0:.4f}'.format(f1))
#이제 f1 스코어도 출력값에 추가
def get_clf_eval(y_test, pred):
confusion = confusion_matrix(y_test, pred)
accuracy = accuracy_score(y_test, pred)
precision = precision_score(y_test, pred)
recall = recall_score(y_test, pred)
#F1 스코어 추가
f1 = f1_score(y_test, pred)
print('오차 행렬')
print(confusion)
#f1 score print 추가
print('정확도: {0:.4f}, 정밀도: {1:.4f}, 재현율: {2:.4f}, F1:{3:.4f}'.format(accuracy, precision, recall, f1))
thresholds = [0.4, 0.45, 0.50, 0.55, 0.60]
pred_proba = lr_clf.predict_proba(X_test)
get_eval_by_threshold(y_test, pred_proba[:,1].reshape(-1, 1), thresholds)[Output]
임계값: 0.4
오차 행렬
[[97 21]
[11 50]]
정확도: 0.8212, 정밀도: 0.7042, 재현율: 0.8197, F1:0.7576
임계값: 0.45
오차 행렬
[[105 13]
[ 13 48]]
정확도: 0.8547, 정밀도: 0.7869, 재현율: 0.7869, F1:0.7869
임계값: 0.5
오차 행렬
[[108 10]
[ 14 47]]
정확도: 0.8659, 정밀도: 0.8246, 재현율: 0.7705, F1:0.7966
임계값: 0.55
오차 행렬
[[111 7]
[ 16 45]]
정확도: 0.8715, 정밀도: 0.8654, 재현율: 0.7377, F1:0.7965
임계값: 0.6
오차 행렬
[[113 5]
[ 17 44]]
정확도: 0.8771, 정밀도: 0.8980, 재현율: 0.7213, F1:0.800005. ROC 곡선과 AUC
ROC 곡선과 이에 기반한 AUC 스코어는 이진 분류의 예측 성능 측정에서 중요하게 사용되는 지표
ROC 곡선은 FPR(False Positive Rate)이 변할 때 TPR(True Positive Rate)이 어떻게 변하는 지 나타내는 곡선
민감도(TPR) 실제값 Positive(양성)가 정확히 예측돼야 하는 수준을 나타냄
특이성(TNR) 실제값 Negative(음성)가 정확히 예측돼야 하는 수준을 나타냄
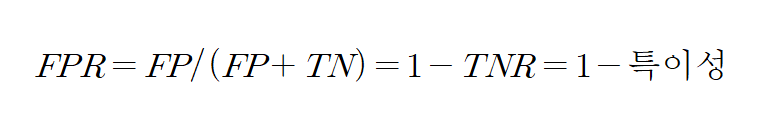
ROC 곡선의 X축 기준 FPR , Y축은 TPR
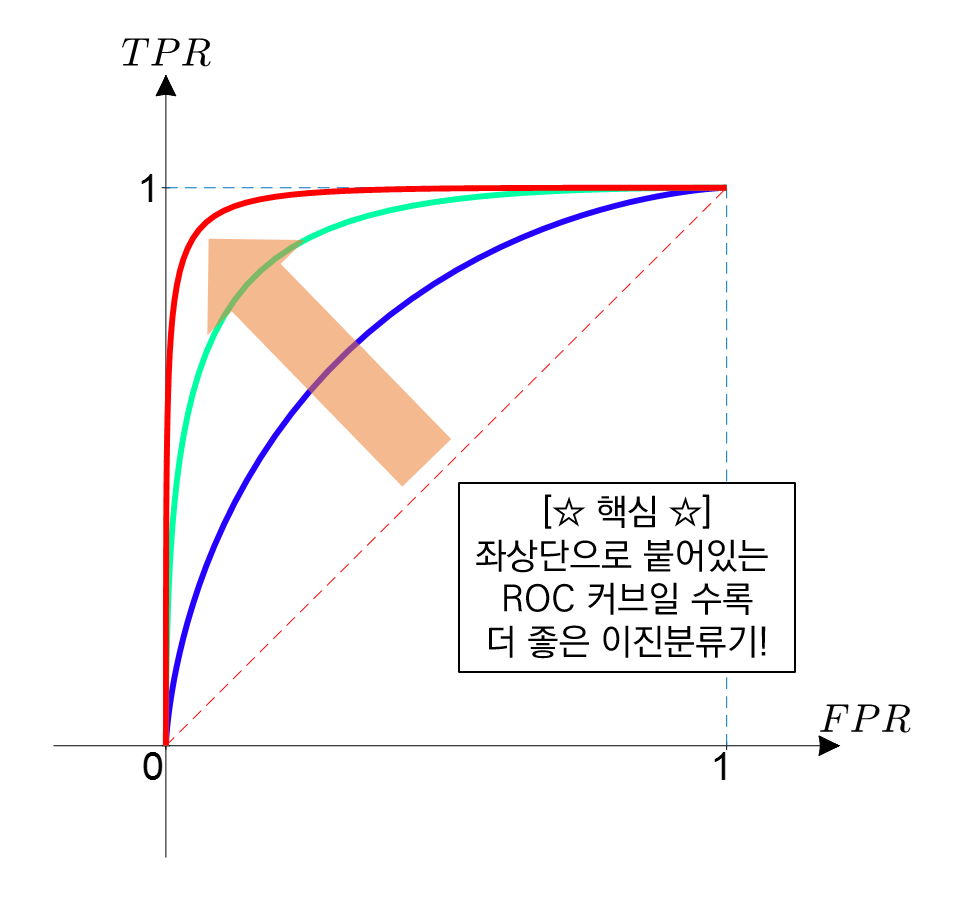
FPR = 0 → 임계값을 1로 지정하면 됨
FPR = 1 → 임계값을 0으로 지정하면 됨
def roc_curve_plot(y_test, pred_proba_c1):
# 임곗값에 따른 FPR, TPR 값을 반환받음.
fprs, tprs, thresholds = roc_curve(y_test, pred_proba_c1)
# ROC 곡선을 그래프 곡선으로 그림
plt.plot(fprs, tprs, label='ROC')
# 가운데 대각선 직선을 그림.
plt.plot([0,1], [0,1], 'k--', label='Random')
# FPR X축의 Scale을 0.1 단위로 변경, X, Y축 명 설정 등
start, end = plt.xlim()
plt.xticks(np.round(np.arange(start, end, 0.1),2))
plt.xlim(0,1)
plt.ylim(0,1)
plt.xlabel('FPR(1 - Specificity)')
plt.ylabel('TPR(Recall)')
plt.legend()
roc_curve_plot(y_test, pred_proba[:,1])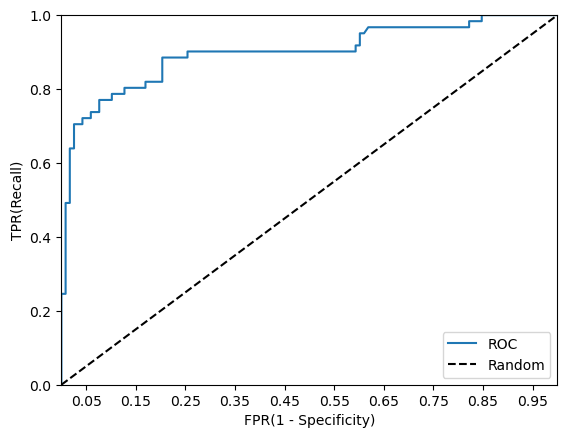
가운데 직선에서 멀어지고 왼쪽 상단 모서리 쪽으로 가파르게 이동할 수록 직사각형에 가까운 곡선이 되어 면적이 1에 가까워지는 좋은 ROC AUC 성능 수치를 얻게 됨
06. 피마 인디언 당뇨병 예측 실습
- 정확도 재현율 보정
#피처 데이터 세트 X, 레이블 데이터 세트 y를 추출
# 맨 끝이 Outcome 칼럼으로 레이블 값임, 칼럼 위치 -1를 이용해 추출
X= diabetes_data.iloc[:,:-1]
y = diabetes_data.iloc[:, -1]
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.2, random_state = 156, stratify=y)
#what is stratify?
# 로지스틱 회귀로 학습, 예측 및 평가 수행
lr_clf = LogisticRegression(solver='liblinear')
lr_clf.fit(X_train, y_train)
pred = lr_clf.predict(X_test)
pred_proba = lr_clf.predict_proba(X_test)[:,1]
get_clf_eval(y_test, pred, pred_proba)[Output]
오차 행렬
[[87 13]
[22 32]]
정확도: 0.7727, 정밀도: 0.7111, 재현율: 0.5926, F1:0.6465, AUC:0.8083전체 데이터의 65%가 Negative이므로 재현율 성능을 올려야됨(재현율 공식에 Negative 있으므로 재현율이 Negative에 초점 맞춘 지표)
- 데이터 전처리
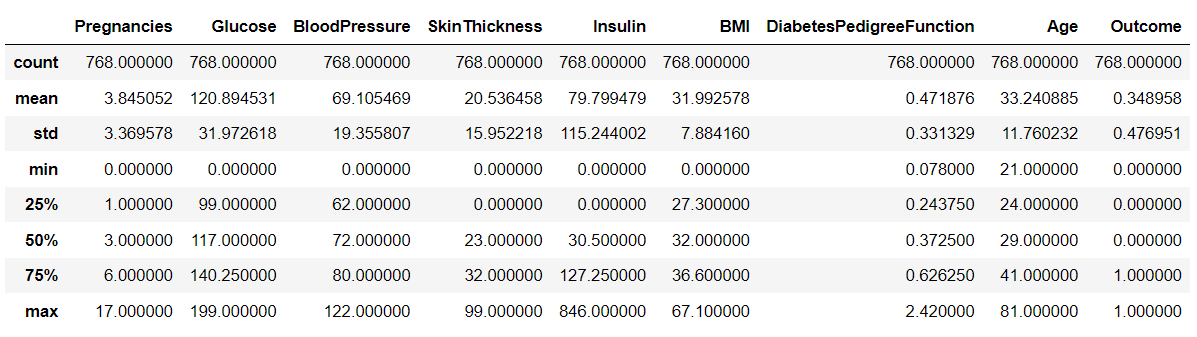
0으로 돼 있는 피처가 많음. 포도당 수치가 0이라는 수치는 잘못된 값.
→ 보정 필요
#0값을 검사할 피처명 리스트
zero_features = ['Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI']
#전체 데이터 건수
total_count = diabetes_data['Glucose'].count()
# 피처별로 반복하면서 데이터 값이 0인 데이터 건수를 추출하고, 퍼센트 계산
for feature in zero_features:
zero_count = diabetes_data[diabetes_data[feature]==0][feature].count()
print('{0} 0 건수는 {1}, 퍼센트는 {2: 2f} %'.format(feature, zero_count, 100*zero_count/total_count))
[Output]
Glucose 0 건수는 5, 퍼센트는 0.651042 %
BloodPressure 0 건수는 35, 퍼센트는 4.557292 %
SkinThickness 0 건수는 227, 퍼센트는 29.557292 %
Insulin 0 건수는 374, 퍼센트는 48.697917 %
BMI 0 건수는 11, 퍼센트는 1.432292 %0 값을 가진 feature가 많기 때문에 0을 평균값으로 대체
# zero_features 리스트 내부에 저장된 개별 피처들에 대하여 0값을 평균 값으로 대체
mean_zero_features = diabetes_data[zero_features].mean()
diabetes_data[zero_features] = diabaetes_data[zero_features].replace(0, mean_zero_features)- 모델 훈련
숫자 데이터에 스케일링 적용
학습/테스트 데이터 세트로 나누기
로지스틱 회귀를 이용해 성능 평가 지표를 확인
# zero_features 리스트 내부에 저장된 개별 피처들에 대해서 0값을 평균 값으로 대체
mean_zero_features = diabetes_data[zero_features].mean()
diabetes_data[zero_features]=diabetes_data[zero_features].replace(0, mean_zero_features)
X= diabetes_data.iloc[:,:-1]
y= diabetes_data.iloc[:,-1]
#StandardScaler 클래스를 이용해 피처 데이터 세트에 일괄적으로 스케일링 적용
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size=0.2, random_state=156, stratify=y)
#로지스틱 회귀로 학습, 예측 및 평가 수행
lr_clf = LogisticRegression()
lr_clf.fit(X_train, y_train)
pred = lr_clf.predict(X_test)
pred_proba = lr_clf.predict_proba(X_test)[:,1]
get_clf_eval(y_test, pred, pred_proba)- 임곗값 찾기
1. 임계값: 0.3
정확도: 0.7143, 정밀도: 0.5658, 재현율: 0.7963
2. 임계값: 0.33
정확도: 0.7403, 정밀도: 0.6000, 재현율: 0.7778
3. 임계값: 0.36
정확도: 0.7468, 정밀도: 0.6190, 재현율: 0.7222
4. 임계값: 0.39
정확도: 0.7532, 정밀도: 0.6333, 재현율: 0.7037
5. 임계값: 0.42
정확도: 0.7792, 정밀도: 0.6923, 재현율: 0.6667
6. 임계값: 0.45
정확도: 0.7857, 정밀도: 0.7059, 재현율: 0.6667
7. 임계값: 0.48
정확도: 0.7987, 정밀도: 0.7447, 재현율: 0.6481
8. 임계값: 0.5
정확도: 0.7987, 정밀도: 0.7674, 재현율: 0.6111임곗값 0.48이 전체적인 성능 평가 지표를 유지하면서 재현율을 약간 향상시키는 좋은 임곗값
- 임곗값 0.48로 훈련 및 예측 클래스 값 도출
#임곗값을 0.48로 설정한 Binarizer 생성
binarizer = Binarizer(threshold=0.48)
#위에서 구한 lr_clf의 predict_proba() 예측 확률 array에서 1에 해당하는 칼럼값을 Binarizer 변환
pred_th_048 = binarizer.fit_transform(pred_proba[:,1].reshape(-1,1))
get_clf_eval(y_test, pred_th_048, pred_proba[:,1])[Output]
오차 행렬
[[88 12]
[19 35]]
정확도: 0.7987, 정밀도: 0.7447, 재현율: 0.6481, F1:0.6931, AUC:0.8433