1. BioPython
Reading FASTA Files
parse(file_path, format)
ex) Counting GC Content in DNA
from Bio.SeqUtils import GCCentral dogma ( DNA --> RNA --> Protein )
Transcription : DNA --> RNA
template_dna = coding_dna.reverse_complement()
messenger_rna = coding_dna.transcribe()
coding_dna = messenger_rna.back_transcribe()Translation : RNA --> Protein
messenger_rna.translate() # *는 종결코돈
coding_dna.translate() # 바로 DNA에서 Protein으로 가능Accessing NCBI databases with Biopython
from Bio import Entrez, SeqIOPairwise Sequence alignments in Biopython
Identify regions of similarity that may indicate functional, structural and evolutionary relationships between two biological sequence. ex) global/local alignment -- > result : match score & gap penalties
from Bio import pairwise2BLAST in Biopython
blast program : blastn, blastp, blastx, tblast, tblastx
from Bio.BLAST import NCBIWWWMotif objects in Biopython
from Bio import motifs
from Bio.Seq import Seq
instances = [
seq("TACAA"),
seq("TACGC")
]
m = motifs.create(instances) # motif로
m.instances # 다시 인스턴스로
m.counts
m.consensus # the largest values in the columns of the .counts matrix
m.anticonsensus # the smallest values in the columns of the .counts matrix
m.degenerate_consensus Indexing a FASTQ file / Sorting a sequence file(FASTA/FASTQ) / Filtering for FASTQ file
참고 : https://www.youtube.com/watch?v=ocA2IMe7dpA
2. RDkit
https://www.rdkit.org/
파이썬에서 약물을 다룰 때 주로 사용하는 package
Drawing Molecules(Jupyter)
from rdkit import Chem
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import Draw
IPythonConsole.ipython_useSVG=True #< set this to False if you want PNGs instead of SVGs
mol = Chem.MolFromSmiles("C1CC2=C3C(=CC=C2)C(=CN3C1)[C@H]4[C@@H](C(=O)NC4=O)C5=CNC6=CC=CC=C65")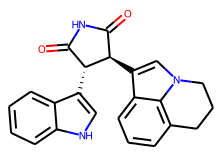
Bonds / Rings / Stereochemistry 분자의 화학적 요소 확인가능
Reactions
rxn = AllChem.ReactionFromSmarts('[cH1:1]1:[c:2](-[CH2:7]-[CH2:8]-[NH2:9]):[c:3]:[c:4]:[c:5]:[c:6]:1.[#6:11]-[CH1;R0:10]=[OD1]>>[c:1]12:[c:2](-[CH2:7]-[CH2:8]-[NH1:9]-[C:10]-2(-[#6:11])):[c:3]:[c:4]:[c:5]:[c:6]:1')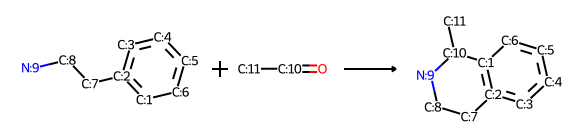
Fingerprint - 분자 내 주요 functional group을 나타냄
Chem.RDKFingerprint() : 2047개로 구분
- 유사도 계산(DTI에서도 자주 사용하지 않을까?)
Tanimoto similarity = 교집합의 갯수 / 합집합의 갯수
DataStructs.FingerprintSimilarity(fps[0],fps[1])
MACCkeys : 166개로 구분
- MACCSkeys.GenMACCSKeys(x)
Morgan : 각 중심 원자에서 얼만큼 떨어진 이웃한 원자까지 고려할지 radius를 정함 --> ECFP
- AllChem.GetMorganFingerprint(x, 2)
Descriptor - 분자의 특성 성질 또는 종합적인 구조/성질을 숫자로 나타내는 것 --> Drug likeness 에 있어서 중요
- TPSA() : Total polar surface area
- MolLogP() : logP 계산
- BalabanJ() : Branch가 얼마나 많은가?
- FractionCSP3() : sp3 carbon 비율
- RingCount() : 고리 갯수
- fr.imide() : imide 존재여부
- fr.phenol() : phenol 존재여부
etc Descriptors 내 method : 425개 / 이와 유사한 rdMolDescriptors(182개)도 존재
참고 : https://www.laidd.org - [2022] RDKit의 기초와 이를 이용한 화학정보학 실습(이주용)
3. PDB-Cleaner
https://github.com/LePingKYXK/PDB_cleaner
Redundant info. in PDB file
--> ligands / alternate location / non-standard a.a residues / negative seq.num / seq. gaps / insertion code / multiple chains / hydrogen atoms
--> REMOVE!! --> SAVE
4. PDBtools
https://github.com/harmslab/pdbtools
For manipulating and doing calculations on wwPDB macromolecule structure files
Functions
1. Structure-based calculations : Geometry / Energy calculation / Structure properties
2. File/structure manipulation
5. PLIP
https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index
 4ASD.PDB
4ASD.PDB
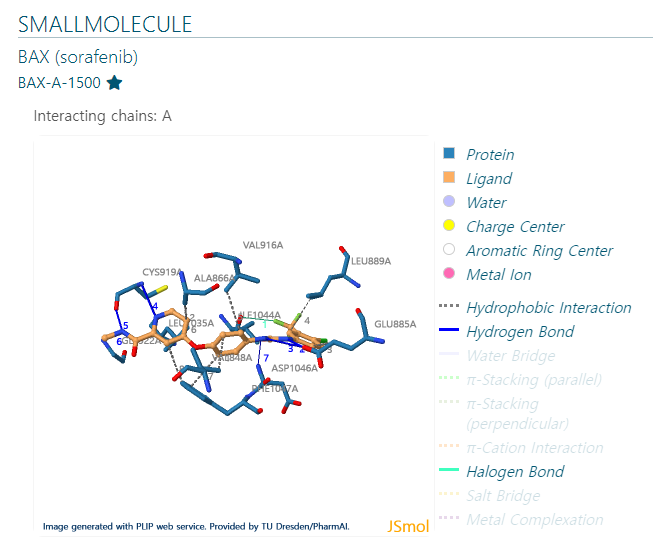
Information of
Hydrophobic Interaction
Hydrogen Bonds
Halogen Bonds
Objective
Main은 small molecule과 protein 간의 interaction을 찾아내는 것.
하지만, PLIP는 nucleic acid(DNA,RNA)의 결합도 찾아낼 수 있다.
- Protein - RNA Interaction
FUS(zinc finger as receptor) <--> RNA(UGGUG as ligand)
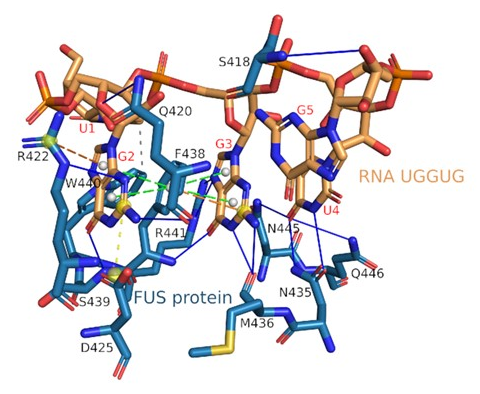
FUS : RNA/DNA binding protein 인 FUS (Fused in sarcoma) 는 신경퇴행성질환인 근위축성 측삭경화증 (Amyothrophic lateral sclerosis)과 전측두엽성치매 (Frontotemproal dementia) 연관 유전자
PLIP detects π-stacking interactions of the central Phe438 with the flanking G2 and G3 of the RNA - S.M. - DNA Interaction
XR5944(cancer drug as ligand) <--> DNA(TFF1-ERE as receptor)
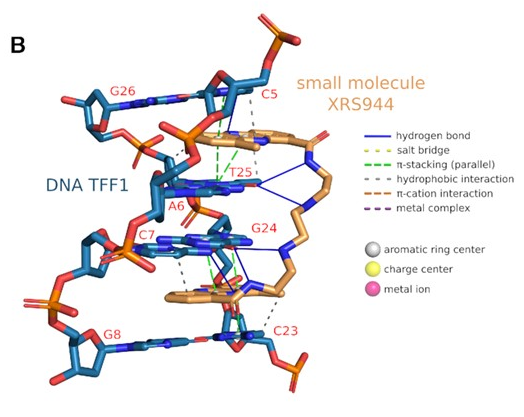 !XR5944 : Estrogen response element(ERE)를 특정하여 인식
!XR5944 : Estrogen response element(ERE)를 특정하여 인식
PLIP identifies parallel π-stacking interactions(phenazine rings of the XR5944 - flanking base) & several hydrophobic interactions at the intercalation sites
