
필요 라이브러리 import
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
import xgboost as xgb
from sklearn import preprocessing
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestRegressor
warnings.simplefilter(action='ignore')데이터 불러오기
Data Dictionary
-
ID : 과수나무 고유 ID
-
착과량(int) : 실제 감귤 착과량 → Target
-
나무 생육 상태 Features (5개)
- 수고(m), 수관폭1(min), 수관폭2(max), 수관폭평균(수관폭1과 수관폭2의 평균)
- 데이터 기입은 cm 단위
-
새순 Features (89개)
- 2022년 09월 01일 ~ 2022년 11월 28일에 일별 측정된 새순 데이터
-
엽록소 Features (89개)
- 2022년 09월 01일 ~ 2022년 11월 28일에 일별 측정된 엽록소 데이터
Train/Test Data
train_data = pd.read_csv("./train.csv")
train_data
test_data = pd.read_csv("./test.csv")
test_data
train_data.info() #데이터 자료형 확인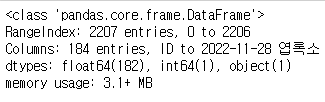
- train_data는 전체 184개의 column이 있으며, 범주형 데이터 1개(ID), int64형 데이터 1개(착과량), float64형 데이터 182개가 존재한다.
train_data.isna().sum().sum() # 결측치 확인
- 전체 데이터에 결측치가 없는 것을 확인할 수 있다.
train_data.describe()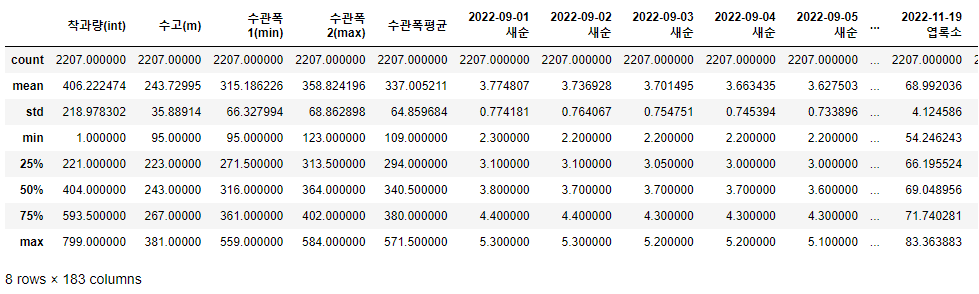
- 전체적인 data가 많아서 한 눈에 보지는 못하지만, 전체적으로 정규분포 형태를 띄고 있으며 크게 outlier가 없는 것도 알 수 있다.
데이터 분석 및 처리
cols = []
for col in train_data.columns:
if '새순' in col or '착과량' in col:
cols.append(col)
sprout = train_data.loc[:, cols]
sprout
cols = []
for col in train_data.columns:
if '엽록' in col or '착과량' in col:
cols.append(col)
chlorophyll = train_data.loc[:, cols]
chlorophyll
- 먼저 착과량과 새순 / 엽록소의 상관관계를 확인하기 위하여 train_data에서 새순 / 엽록소, 착과량 부분을 추출하여 dataframe을 만들었다.
plt.figure(figsize = (12, 8))
x = sns.heatmap(sprout.corr(), cmap = 'Blues', linewidths = '0.1',annot = False)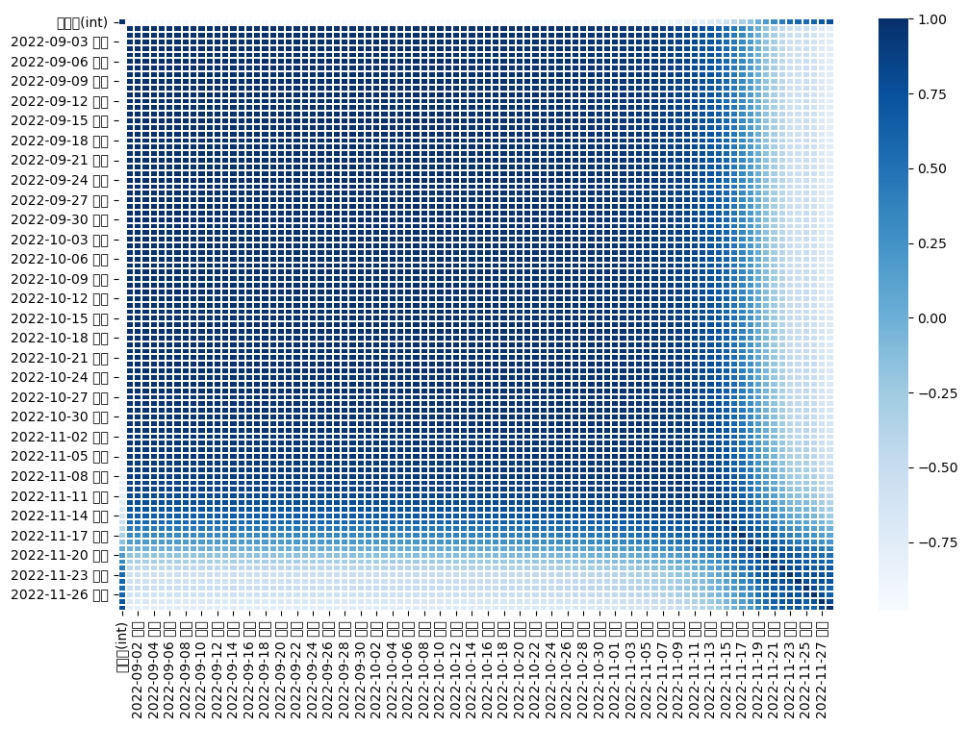
plt.figure(figsize = (12, 8))
x = sns.heatmap(chlorophyll.corr(), cmap = 'Blues', linewidths = '0.1',annot = False)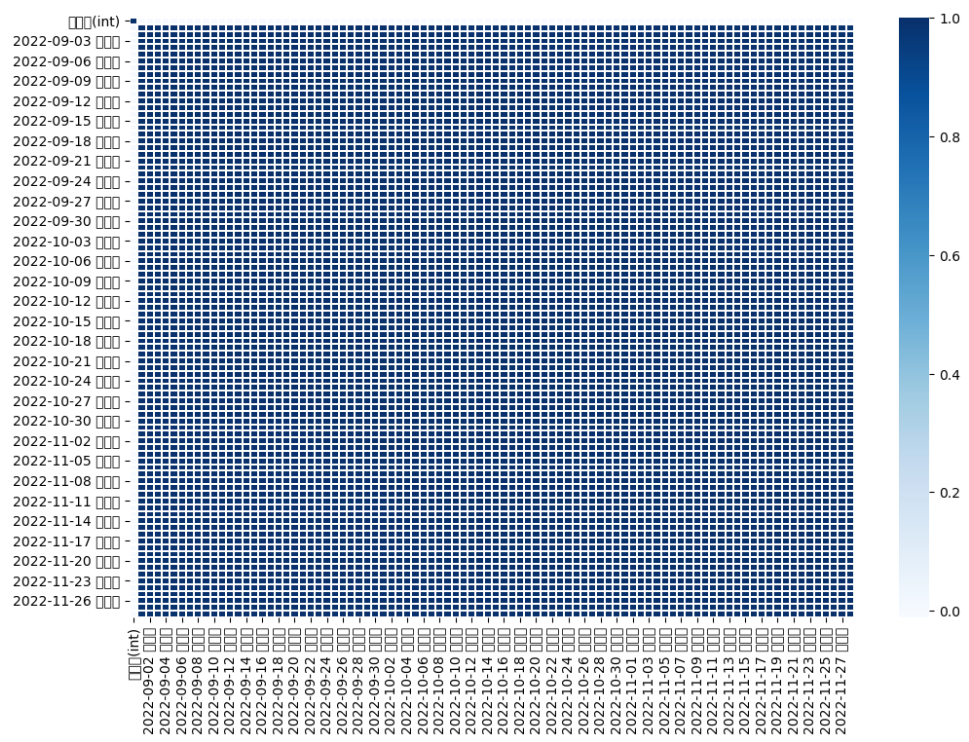
- 새순과 엽록소, 착과량의 상관관계를 확인해 본 결과 11월달의 새순 데이터만 착과량과 상관관계가 유의미한 것을 볼 수 있었다.
train_data['새순1'] = train_data.iloc[:, 6:11].mean(axis=1)
train_data['새순2'] = train_data.iloc[:, 11:16].mean(axis=1)
train_data['새순3'] = train_data.iloc[:, 16:21].mean(axis=1)
train_data['새순4'] = train_data.iloc[:, 21:26].mean(axis=1)
train_data['새순5'] = train_data.iloc[:, 26:31].mean(axis=1)
train_data['새순6'] = train_data.iloc[:, 31:36].mean(axis=1)
train_data['새순7'] = train_data.iloc[:, 36:41].mean(axis=1)
train_data['새순8'] = train_data.iloc[:, 41:46].mean(axis=1)
train_data['새순9'] = train_data.iloc[:, 46:51].mean(axis=1)
train_data['새순10'] = train_data.iloc[:, 51:56].mean(axis=1)
train_data['새순11'] = train_data.iloc[:, 56:61].mean(axis=1)
train_data['새순12'] = train_data.iloc[:, 61:66].mean(axis=1)
train_data['새순13'] = train_data.iloc[:, 66:71].mean(axis=1)
train_data['새순14'] = train_data.iloc[:, 71:76].mean(axis=1)
train_data['새순15'] = train_data.iloc[:, 76:81].mean(axis=1)
train_data['새순16'] = train_data.iloc[:, 81:86].mean(axis=1)
train_data['새순17'] = train_data.iloc[:, 86:91].mean(axis=1)
train_data['새순18'] = train_data.iloc[:, 91:95].mean(axis=1)
train_data = train_data.drop(train_data.columns[6:95], axis=1)train_data['엽록소1'] = train_data.iloc[:, 6:11].mean(axis=1)
train_data['엽록소2'] = train_data.iloc[:, 11:16].mean(axis=1)
train_data['엽록소3'] = train_data.iloc[:, 16:21].mean(axis=1)
train_data['엽록소4'] = train_data.iloc[:, 21:26].mean(axis=1)
train_data['엽록소5'] = train_data.iloc[:, 26:31].mean(axis=1)
train_data['엽록소6'] = train_data.iloc[:, 31:36].mean(axis=1)
train_data['엽록소7'] = train_data.iloc[:, 36:41].mean(axis=1)
train_data['엽록소8'] = train_data.iloc[:, 41:46].mean(axis=1)
train_data['엽록소9'] = train_data.iloc[:, 46:51].mean(axis=1)
train_data['엽록소10'] = train_data.iloc[:, 51:56].mean(axis=1)
train_data['엽록소11'] = train_data.iloc[:, 56:61].mean(axis=1)
train_data['엽록소12'] = train_data.iloc[:, 61:66].mean(axis=1)
train_data['엽록소13'] = train_data.iloc[:, 66:71].mean(axis=1)
train_data['엽록소14'] = train_data.iloc[:, 71:76].mean(axis=1)
train_data['엽록소15'] = train_data.iloc[:, 76:81].mean(axis=1)
train_data['엽록소16'] = train_data.iloc[:, 81:86].mean(axis=1)
train_data['엽록소17'] = train_data.iloc[:, 86:91].mean(axis=1)
train_data['엽록소18'] = train_data.iloc[:, 91:95].mean(axis=1)
train_data = train_data.drop(train_data.columns[6:95], axis=1)
train_data
- 새순과 엽록소가 각 89개의 데이터가 있는데, 이러한 데이터를 그대로 모델링을 한다면 overfitting이 일어날 수도 있고, 각 날짜별로 차이가 안나는 데이터도 존재하기 때문에 5일 단위로 묶어 평균치로 새로운 column을 만들고 기존 데이터는 drop하였다.
train_data['수관폭 Max-Min'] = train_data.iloc[:, 4] - train_data.iloc[:, 3]
train_data = train_data.drop(['ID', '수관폭1(min)', '수관폭2(max)'], axis = 1)
train_data
- 수관 폭의 평균과 함께 최대-최소 값이 유용할 수 있어서 해당 column을 하나 생성하고, 기준의 수관폭 max, min과 쓸모 없는 ID column을 drop하였다.
scaler = preprocessing.MinMaxScaler()
normalized_data = scaler.fit_transform(train_data)
col = ['착과량(int)', '수고(m)', '수관폭평균', '새순1', '새순2', '새순3', '새순4', '새순5', '새순6',
'새순7', '새순8', '새순9', '새순10', '새순11', '새순12', '새순13', '새순14', '새순15',
'새순16', '새순17', '새순18', '엽록소1', '엽록소2', '엽록소3', '엽록소4', '엽록소5', '엽록소6',
'엽록소7', '엽록소8', '엽록소9', '엽록소10', '엽록소11', '엽록소12', '엽록소13', '엽록소14',
'엽록소15', '엽록소16', '엽록소17', '엽록소18', '수관폭 Max-Min']
normalized_train = pd.DataFrame(normalized_data, columns = col)
normalized_train
- 각 column들의 단위가 다르고, 특정 column에 가중치가 생기는 것을 방지하기 위하여 MinMaxScaler를 통하여 데이터 전처리를 진행하였다.
Modeling
y_target = train_data['착과량(int)']
x_data = normalized_train.drop(['착과량(int)'], axis = 1)
x_train, x_test, y_train, y_test = train_test_split(x_data, y_target, test_size = 0.2, random_state=42)
print(x_train.shape, x_test.shape, y_train.shape, y_test.shape)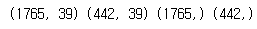
- target과 data로 나눈 뒤 이를 train_test_split으로 test용 data와 train용 data로 분리를 한 뒤 확인을 하였다.
RandomForestRegressor
params ={
'n_estimators':[50, 100, 200, 300, 400, 500],
'max_depth':[6, 8, 10, 12, 16],
'min_samples_leaf':[4, 8, 12, 16],
'min_samples_split':[4, 8, 12, 16, 20]
}
rfr = RandomForestRegressor()
grid_cv = GridSearchCV(rfr, param_grid=params, cv=2, n_jobs=-1)
grid_cv.fit(x_train,y_train)
print('최적 하이퍼 파라미터: ', grid_cv.best_params_)
print('최고 예측 정확도: {:.4f}'.format(grid_cv.best_score_))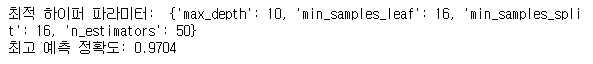
# 모델 학습
model = RandomForestRegressor(n_estimators = 50, max_depth = 10, min_samples_leaf = 16, min_samples_split = 16, random_state=42, oob_score=True)
model.fit(x_train, y_train)
pred = model.predict(x_test)
# 평가
print("훈련 세트 정확도: {:.3f}".format(model.score(x_train, y_train)) )
print("테스트 세트 정확도: {:.3f}".format(model.score(x_test, y_test)) )
print("OOB 샘플의 정확도: {:.3f}".format(model.oob_score_) )
- GridSearchCV를 이용하여 RandomForestRegressor 모델의 최적 하이퍼 파라미터 튜닝을 진행한 다음 모델링을 진행하였다.
XGBoostRegressor
params ={
'n_estimators':[50, 100, 200, 300, 400, 500],
'learning_rate':[0.1, 0.2, 0.3],
'gamma':[0, 1, 2],
'subsample':[0.3, 0.5, 0.7],
'max_depth':[4, 5, 6, 7, 8]
}
xgbr = xgb.XGBRegressor()
grid_cv = GridSearchCV(xgbr, param_grid=params, cv=2, n_jobs=-1)
grid_cv.fit(x_train,y_train)
print('최적 하이퍼 파라미터: ', grid_cv.best_params_)
print('최고 예측 정확도: {:.4f}'.format(grid_cv.best_score_))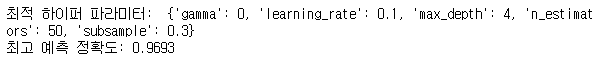
# 모델 학습
xbgmodel = xgb.XGBRegressor(n_estimators=50, learning_rate=0.1, gamma=0, subsample=0.75, max_depth=4, random_state=42)
xgbmodel.fit(x_train, y_train)
pred = model.predict(x_test)
# 평가
print("훈련 세트 정확도: {:.3f}".format(xgbmodel.score(x_train, y_train)) )
print("테스트 세트 정확도: {:.3f}".format(xgbmodel.score(x_test, y_test)) )
- GridSearchCV를 이용하여 XGBoostRegressor 모델의 최적 하이퍼 파라미터 튜닝을 진행한 다음 모델링을 진행하였다.
