코드스테이츠 AI Bootcamp Section2에서 자유주제로 진행한 Machine Learning 개인프로젝트 내용 정리 및 회고. (3) Model 해석, 결론, 회고
- (1) 서론과 데이터준비 과정
- (2) 모델링 및 모델 하이퍼파라미터 튜닝
- (3) Model 해석, 결론, 회고
- Github Repository 바로가기 : Project-Hypertension-Predictive-Model (클릭)
한국형 고혈압 예측 모델 개발 (Hypertension Predictive Model)
- 고혈압 진단기준에 따른 차이가 있을까?

- Tech Stack

00. 프로젝트 개요
필수 포함 요소
자유 주제로 직접 선택한 데이터셋을 사용머신러닝 예측 모델을 통한성능 및 인사이트를 도출/공유- 데이터셋
전처리/EDA부터모델을 해석하는 과정까지 수행 - 발표를 듣는 사람은
비데이터 직군이라 가정
프로젝트 목차
-
Introduction
서론- Hypertension
고혈압 - Diagnotic Criteria
고혈압 진단기준 - Problems & Goals
문제정의 및 목표 - Dataset
선정 데이터셋 - Target & Features
타겟과 특성
- Hypertension
-
Data Preparation
데이터 준비- Data Pre-processing
데이터 전처리 - Final Data
최종데이터
- Data Pre-processing
-
Modeling
모델링- Data split
데이터 분리 - Baseline & Metrics
기준모델 및 평가지표 - Modeling(default)
기본값으로 모델링 - Hyperparameter Tuning
모델 튜닝 - Final Model & Test
최종모델 및 테스트
- Data split
-
Interpretation
해석- Feature Importance (MDI)
특성중요도 - Permutation Importance
순열중요도 - Feature Analysis (PDP)
특성 분석
- Feature Importance (MDI)
-
Conclusion
결론- Summary
요약 - Conclusion & Limitation
결론 및 한계점 - Retrospective
회고
- Summary
04. Interpretation 해석
4-0. Library & Data Import
- 튜닝과정이 오래걸려서 해석부분부터 새로 파일을 만들어 진행한 관계로 다시 라이브러리와 데이터를 불러와서 진행
Library import
import pandas as pd
from pandas_profiling import ProfileReport
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
plt.rcParams['font.family'] = 'Malgun Gothic'
plt.rcParams['axes.unicode_minus'] = False
import seaborn as sns
#모델 학습 및 검증점수 일치여부 확인을 위한 라이브러리
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, roc_auc_score, roc_curve, confusion_matrix
from xgboost import XGBClassifier
from sklearn.model_selection import cross_val_score, cross_val_predict, KFold
#모델 해석을 위한 라이브러리
import eli5
from eli5.sklearn import PermutationImportance
from pdpbox.pdp import pdp_isolate, pdp_plot, pdp_interact, pdp_interact_plotData import
df0 = pd.read_csv('data/KNHANES_8th_final2.csv')
df1 = df0.iloc[:,2:]
df1.info()
#output
'''
RangeIndex: 11422 entries, 0 to 11421
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 sex 11422 non-null int64
1 age 11422 non-null int64
2 heavy_drink 11422 non-null int64
3 smoke 11422 non-null int64
4 genetic_hbp 11422 non-null float64
5 BMI 11422 non-null float64
6 diabetes 11422 non-null int64
7 hyper_chol 11422 non-null int64
8 triglycerides 11422 non-null float64
9 HBP_US 11422 non-null int64
10 HBP_EU 11422 non-null int64
dtypes: float64(3), int64(8)
memory usage: 981.7 KB
'''최종모델 학습
- 최종모델 : 튜닝과정을 거친 XGBoost model
- 학습과정 및 검증은 이전 포스팅과 동일하므로 생략
테스트 데이터셋 특성상관관계 확인
- 특성간 상관관계가 큰 관계가 존재하면 해석에 유의해야하므로 확인하였음.
- 확인결과 특성간 상관관계가 크지 않았음
- 상관 관계를 고려하지 않고 해석하도록 하겠음.
corr_test = X_test.corr()
fig, ax = plt.subplots(figsize=(12,8))
mask = np.zeros_like(corr_test,dtype=bool)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(corr_test,
cmap='coolwarm',
annot=True,
mask=mask,
linewidths=.5,
cbar_kws={'shrink':.5},
vmin=-1,
vmax=1)
plt.title('Correlation of features (X_test)')
plt.show()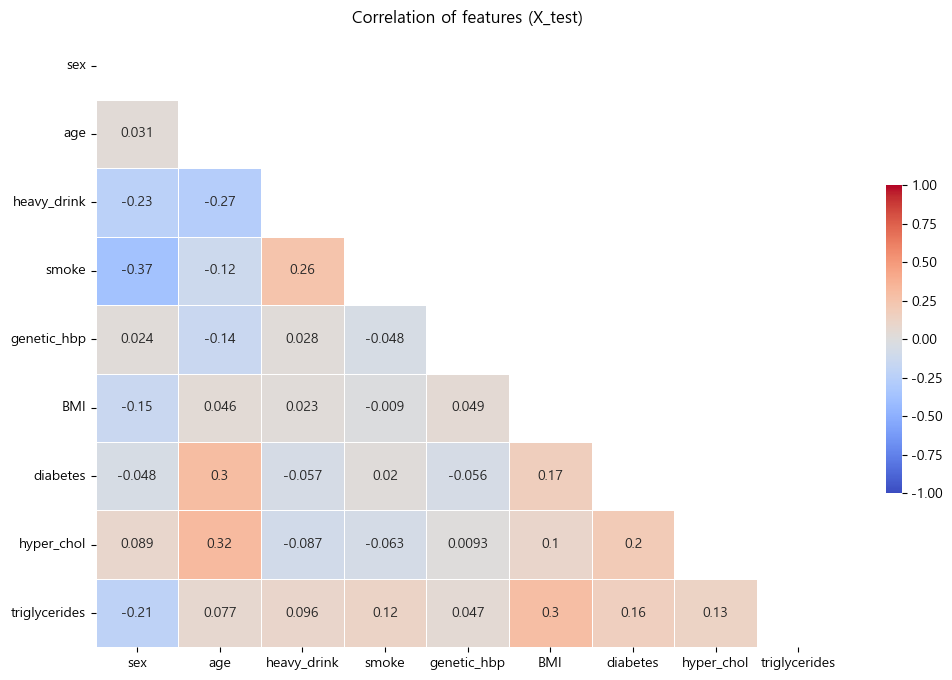
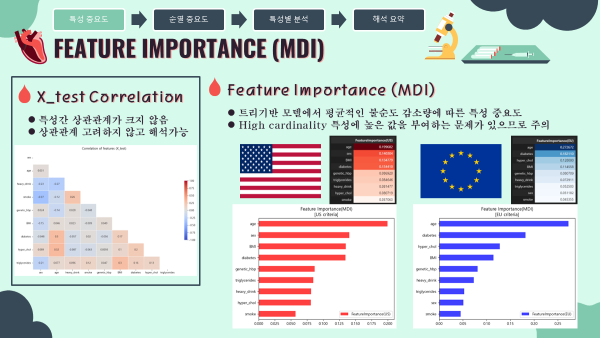
4-1. Feature Importance (MDI) 특성중요도
- MDI 특성중요도는 트리기반 모델에서 평균적인 불순도 감소량에 따른 특성 중요도를 나타냄
- High cardinality(다양성) 특성에 높은 값을 부여하는 문제가 있으므로 이 다음에 실시할 순열중요도를 더 중점적으로 해석하였음.
미국 진단기준 (130mmHg/80mmHg)
importances_us = pd.DataFrame(data={'FeatureImportance(US)':xgb_us_tun.feature_importances_},index=X_train.columns)
display(importances_us.sort_values(by='FeatureImportance(US)',ascending=False).style.background_gradient(cmap='Reds'))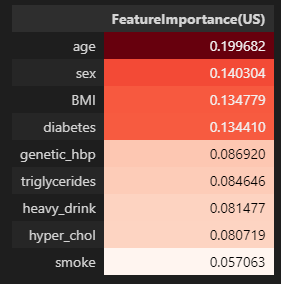
importances_us.sort_values(by='FeatureImportance(US)').plot.barh(color='red',alpha=0.75)
plt.title('Feature Importance(MDI)\n[US criteria]')
plt.legend(loc='lower right')
plt.show()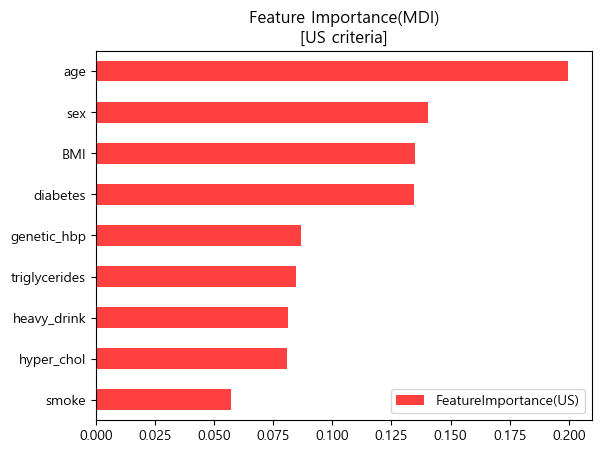
유럽 진단기준 (140mmHg/90mmHg)
importances_eu = pd.DataFrame(data={'FeatureImportance(EU)':xgb_eu_tun.feature_importances_},index=X_train.columns)
display(importances_eu.sort_values(by='FeatureImportance(EU)',ascending=False).style.background_gradient(cmap='Blues'))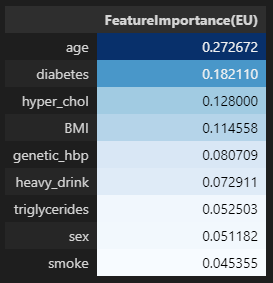
importances_eu.sort_values(by='FeatureImportance(EU)').plot.barh(color='blue',alpha=0.75)
plt.title('Feature Importance(MDI)\n[EU criteria]')
plt.legend(loc='lower right')
plt.show()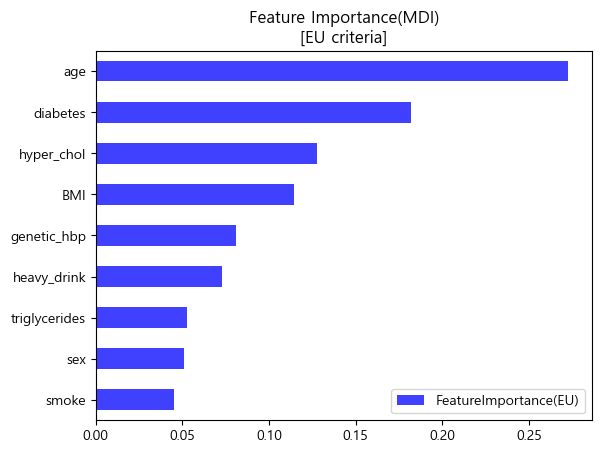
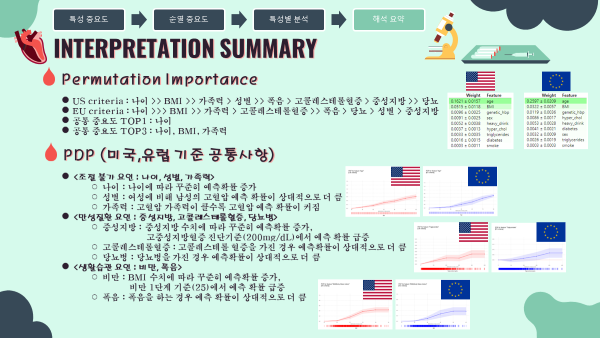
4-2. Permutation Importance 순열중요도
- 순열중요도란 하나의 특성에 노이즈를 주어서 얼마나 성능이 변하는 지를 살펴보는 방법
- MDI 특성중요도의 단점을 보완한 특성의 중요도 산출 방법
- MDI보다 High Cardinality특성에 덜 치우진 결과를 보여줌
- 모든 모델에 범용적으로 적용 가능한 Model-Agnostic한 방법
- MDI 특성중요도의 단점을 보완한 특성의 중요도 산출 방법
- 미국과 유럽 공통적으로 나이, 비만도(BMI), 가족력을 가장 중요한 특성이라 판단하였음.
- 0에 거의 수렴하는 흡연여부는 이후 분석에서 제외하고 해석을 진행하였음.
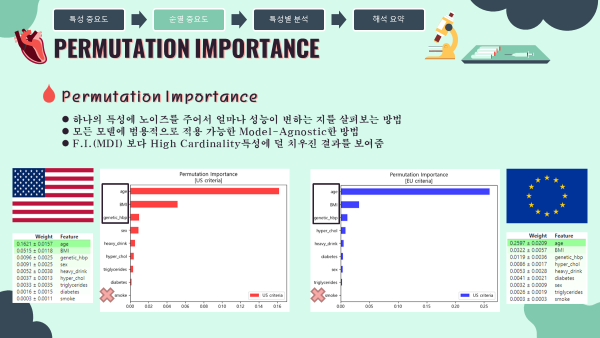
미국 진단기준 (130mmHg/80mmHg)
- Permutation Importance fit (eli5.sklearn)
permuter_us = PermutationImportance(
xgb_us_tun,
scoring='roc_auc',
n_iter=10,
random_state=42
)
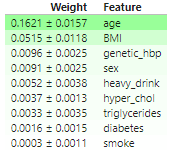
permuter_us.fit(X_test,y_test_us)- 가중치 DataFrame (eli5)
features_us = X_test.columns.to_list()
display(
eli5.show_weights(
permuter_us,
top=None,
feature_names=features_us
))
- 순열중요도 barplot (matplotlib)
pi_us = pd.Series(permuter_us.feature_importances_,features_us).sort_values()
pi_us.plot.barh(color='red',alpha=0.75,label='US criteria')
plt.legend(loc='lower right')
plt.title('Permutation Importance\n[US criteria]')
plt.show()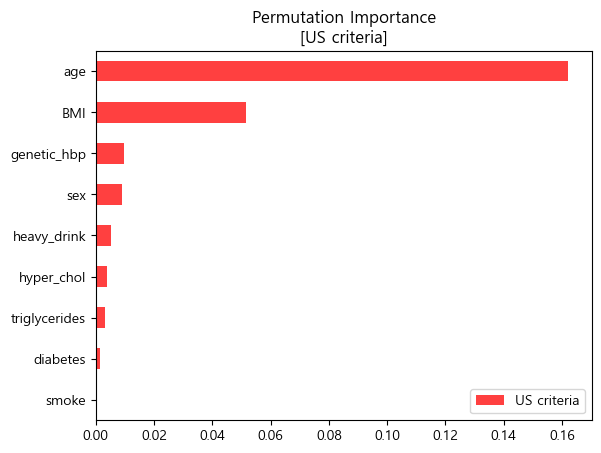
유럽 진단기준 (140mmHg/90mmHg)
- Permutation Importance fit (eli5.sklearn)
permuter_eu = PermutationImportance(
xgb_eu_tun,
scoring='roc_auc',
n_iter=10,
random_state=42
)
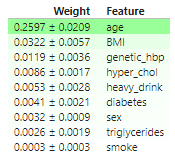
permuter_eu.fit(X_test,y_test_eu)- 가중치 DataFrame (eli5)
features_eu = X_test.columns.to_list()
display(
eli5.show_weights(
permuter_eu,
top=None,
feature_names=features_eu
))
- 순열중요도 barplot (matplotlib)
pi_eu = pd.Series(permuter_eu.feature_importances_,features_eu).sort_values()
pi_eu.plot.barh(color='blue',alpha=0.75,label='US criteria')
plt.legend(loc='lower right')
plt.title('Permutation Importance\n[EU criteria]')
plt.show()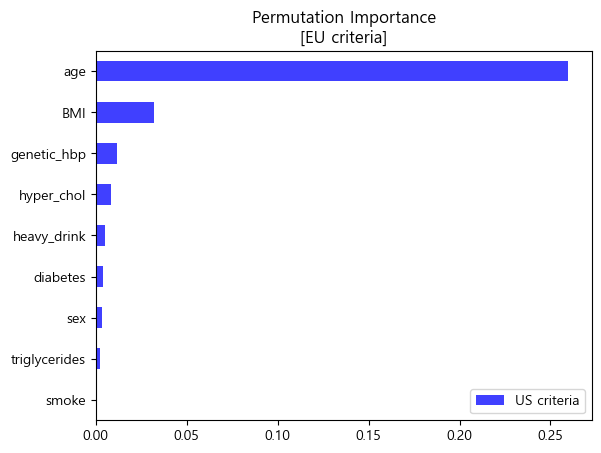
4-3. Feature Analysis (PDP) 특성 분석
- 각 특성에 따른 경향성을 파악하기 위해 PDP(Partial Dependence Plot)을 진행함.
- PDP란 평균적으로 모델이 특정 특성의 변화에 따라 어떻게 예측하는지를 보여주는 그래프.
- 각 특성간의 독립성을 전제로 하기 때문에, 특성간 상관관계가 있는 경우 해석에 유의해야함.
- 본 프로젝트에선 특성간 상관관계가 크지 않기 때문에 해석에는 지장이 없을 것으로 판단함.
조절 불가 요인 (나이, 성별, 가족력)
- 나이 : 연령이 높을 수록 고혈압 예측 확률이 증가하는 경향을 보임
- 성별 : 여성에비해 남성이 더 높은 고혈압 확률을 나타냄
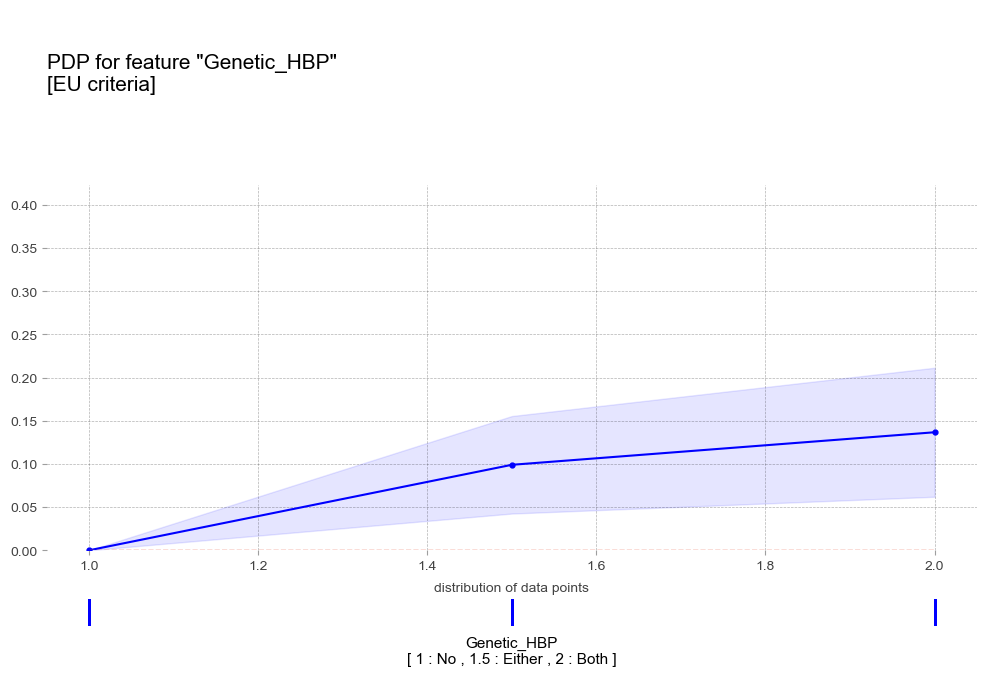
- 가족력 : 가족력이 강할수록 고혈압 예측 확률이 증가하는 경향을 보임
- 나이 Age [연속형변수]
pdp_age_us = pdp_isolate(
model=xgb_us_tun,
dataset=X_test,
model_features=X_test.columns,
feature='age',
grid_type='percentile',
cust_grid_points=[20,30,40,50,60,70,80]
)
pdp_plot(pdp_age_us,
'Age',
plot_pts_dist=True,
figsize=(12,8),
plot_params={
'title':'PDP for feature "Age"\n[US criteria]',
'subtitle':'',
'fill_color':'red',
'fill_alpha':0.1,
'pdp_color':'red'
})
plt.show()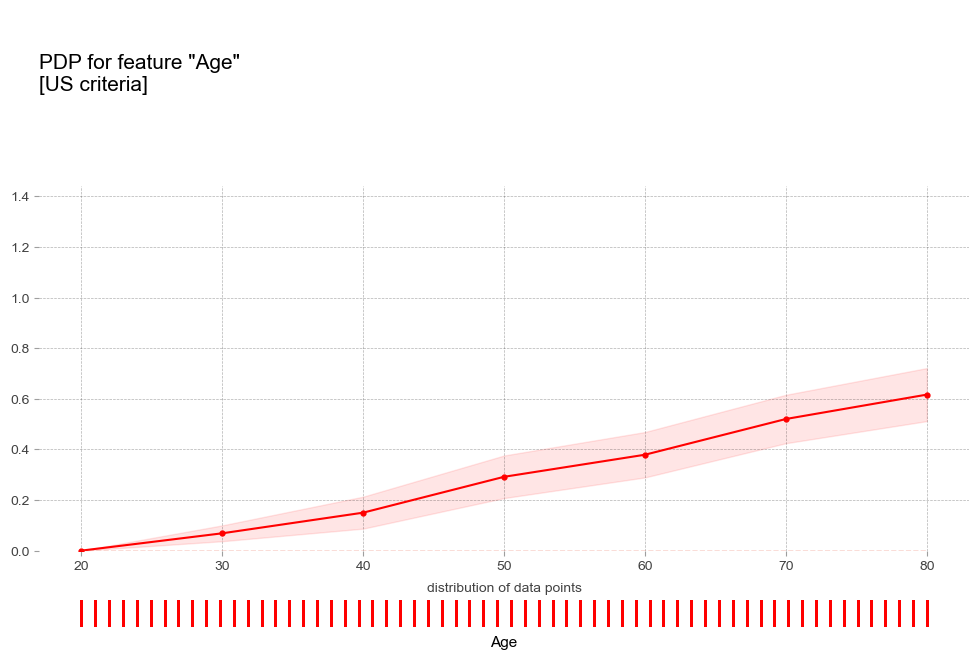
pdp_age_eu = pdp_isolate(
model=xgb_eu_tun,
dataset=X_test,
model_features=X_test.columns,
feature='age',
grid_type='percentile',
cust_grid_points=[20,30,40,50,60,70,80]
)
pdp_plot(pdp_age_eu,
'Age',
plot_pts_dist=True,
figsize=(12,8),
plot_params={
'title':'PDP for feature "Age"\n[EU criteria]',
'subtitle':'',
'fill_color':'blue',
'fill_alpha':0.1,
'pdp_color':'blue'
})
plt.show()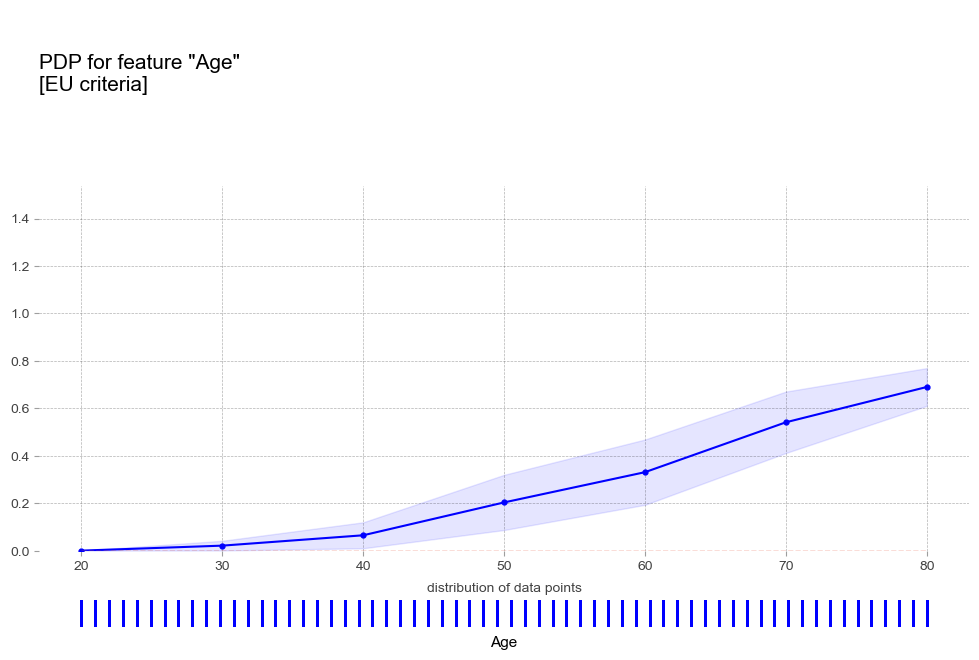
- 성별 Sex [범주형변수]
pdp_sex_us = pdp_isolate(
model=xgb_us_tun,
dataset=X_test,
model_features=X_test.columns,
feature='sex',
grid_type='percentile',
cust_grid_points=[1,2]
)
pdp_plot(pdp_sex_us,
'Sex\n[ 1 : Man , 2 : Woman ]',
figsize=(12,8),
plot_params={
'title':'PDP for feature "Sex"\n[US criteria]',
'subtitle':'',
'fill_color':'red',
'fill_alpha':0.1,
'pdp_color':'red'
})
plt.show()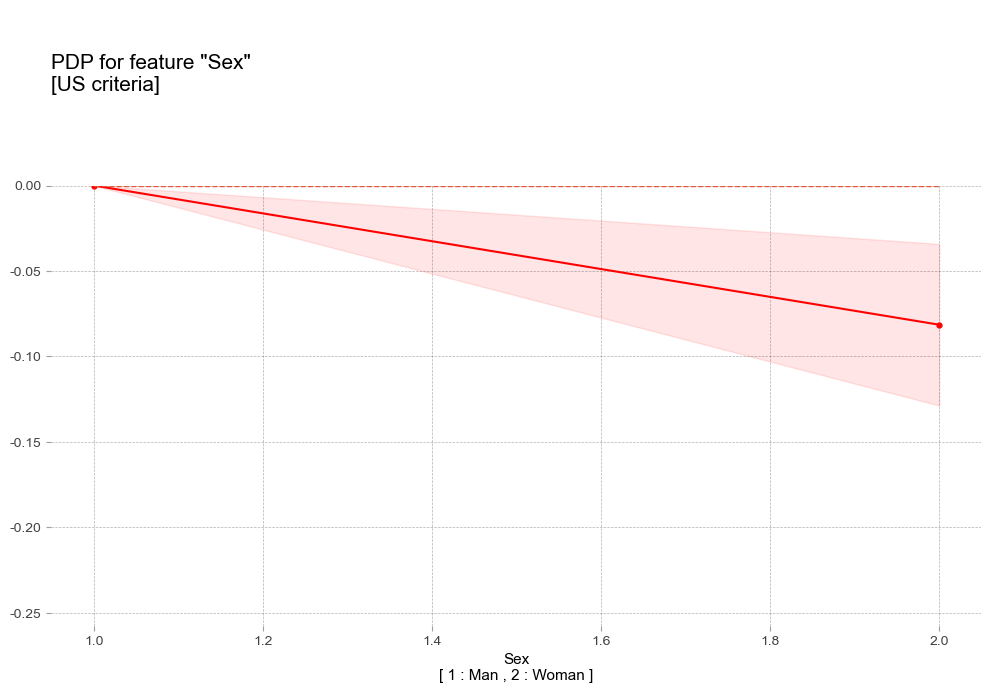
pdp_sex_eu = pdp_isolate(
model=xgb_eu_tun,
dataset=X_test,
model_features=X_test.columns,
feature='sex',
grid_type='percentile',
cust_grid_points=[1,2]
)
pdp_plot(pdp_sex_eu,
'Sex\n[ 1 : Man , 2 : Woman ]',
plot_pts_dist=True,
figsize=(12,8),
plot_params={
'title':'PDP for feature "Sex"\n[EU criteria]',
'subtitle':'',
'fill_color':'blue',
'fill_alpha':0.1,
'pdp_color':'blue'
})
plt.show()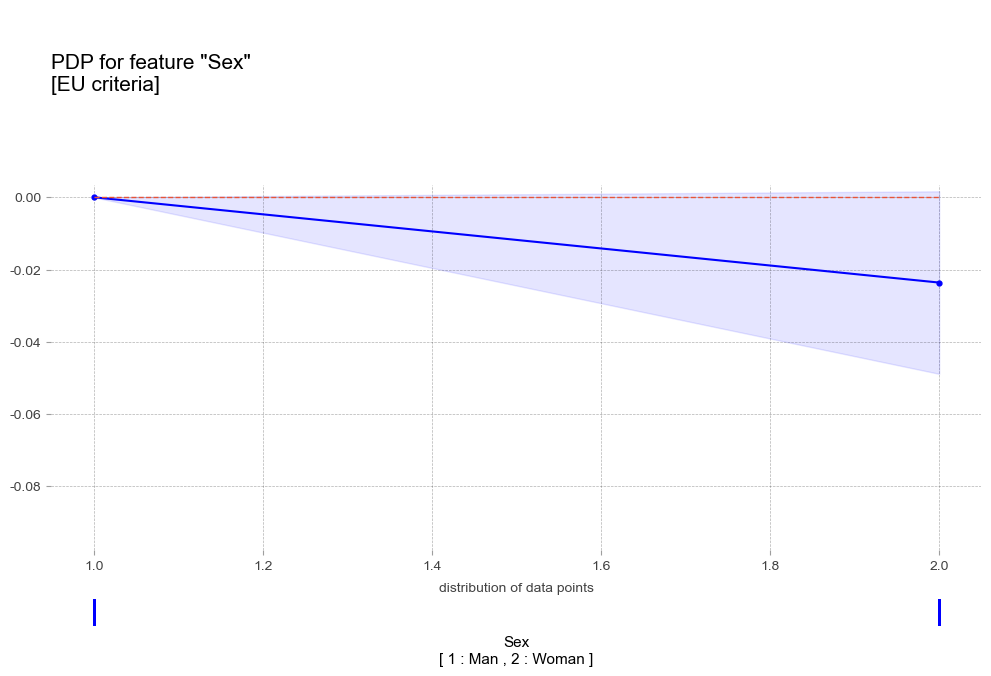
- 가족력 Genetic(HBP) [범주형변수]
pdp_gen_us = pdp_isolate(
model=xgb_us_tun,
dataset=X_test,
model_features=X_test.columns,
feature='genetic_hbp',
grid_type='percentile',
cust_grid_points=[1,1.5,2]
)
pdp_plot(pdp_gen_us,
'Genetic_HBP\n[ 1 : No , 1.5 : Either , 2 : Both ]',
plot_pts_dist=True,
figsize=(12,8),
plot_params={
'title':'PDP for feature "Genetic_HBP"\n[US criteria]',
'subtitle':'',
'fill_color':'red',
'fill_alpha':0.1,
'pdp_color':'red'
})
plt.show()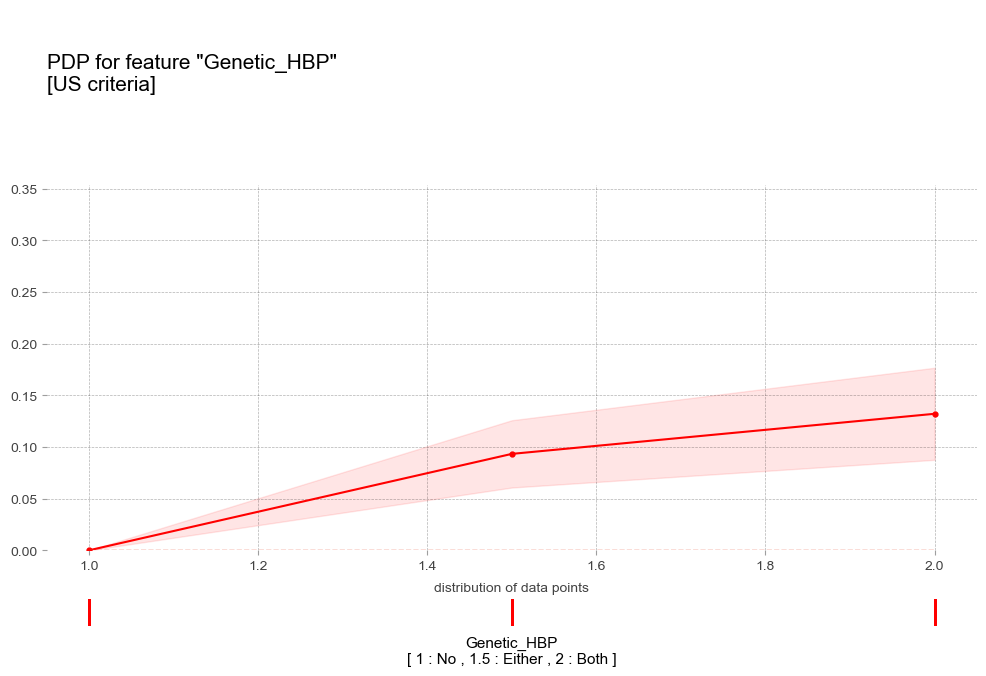
pdp_gen_eu = pdp_isolate(
model=xgb_eu_tun,
dataset=X_test,
model_features=X_test.columns,
feature='genetic_hbp',
grid_type='percentile',
cust_grid_points=[1,1.5,2]
)
pdp_plot(pdp_gen_eu,
'Genetic_HBP\n[ 1 : No , 1.5 : Either , 2 : Both ]',
plot_pts_dist=True,
figsize=(12,8),
plot_params={
'title':'PDP for feature "Genetic_HBP"\n[EU criteria]',
'subtitle':'',
'fill_color':'blue',
'fill_alpha':0.1,
'pdp_color':'blue'
})
plt.show()
만성질환 요인 (중성지방, 고콜레스테롤혈증, 당뇨병)
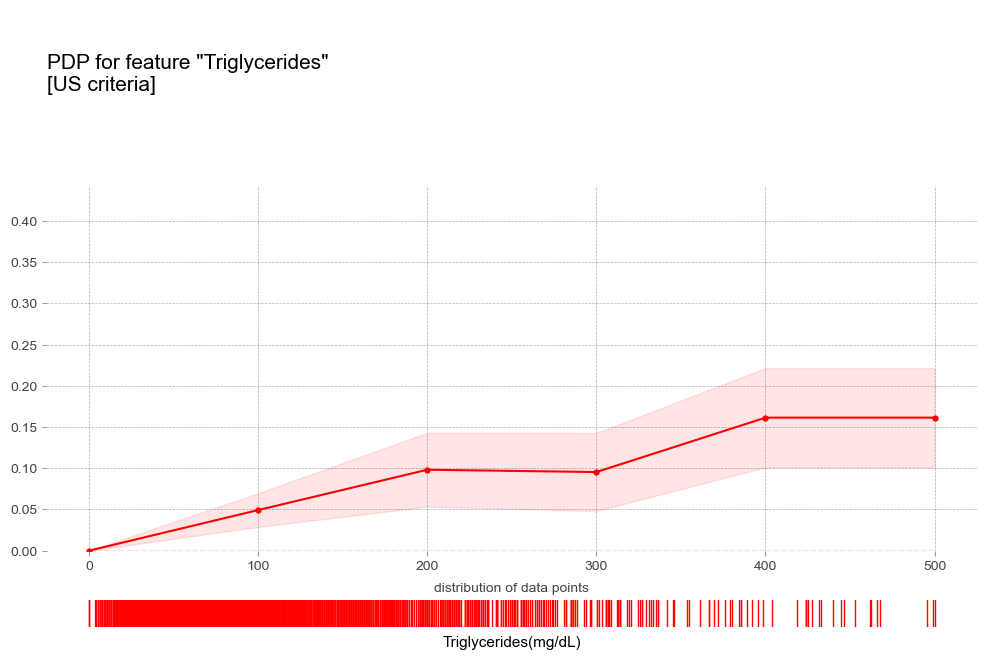
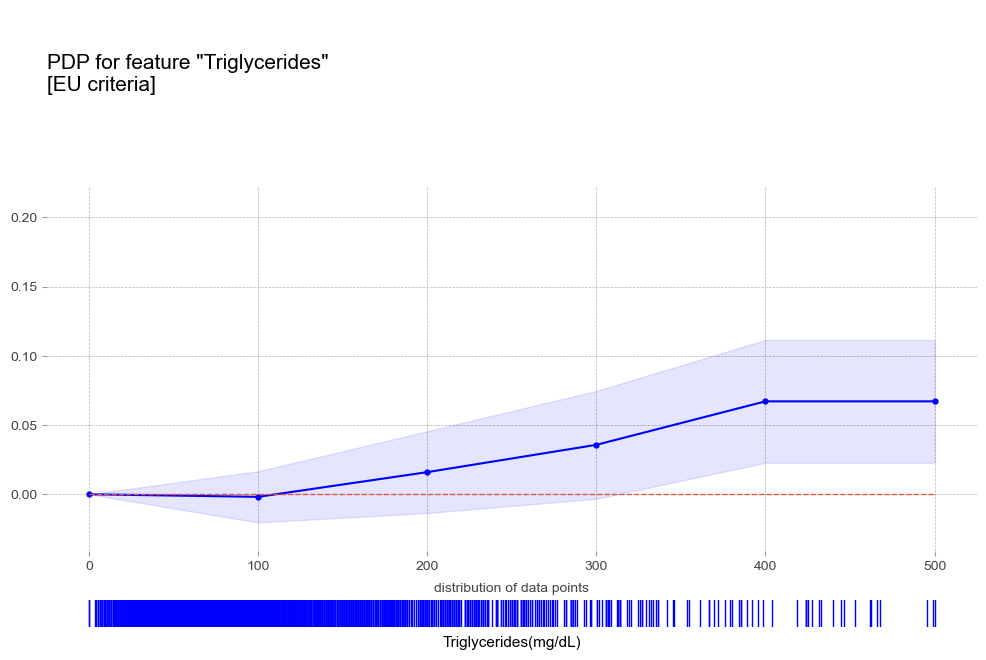
- 중성지방 : 중성지방 수치가 높을 수록 고혈압 예측 확률이 증가하는 경향을 보임
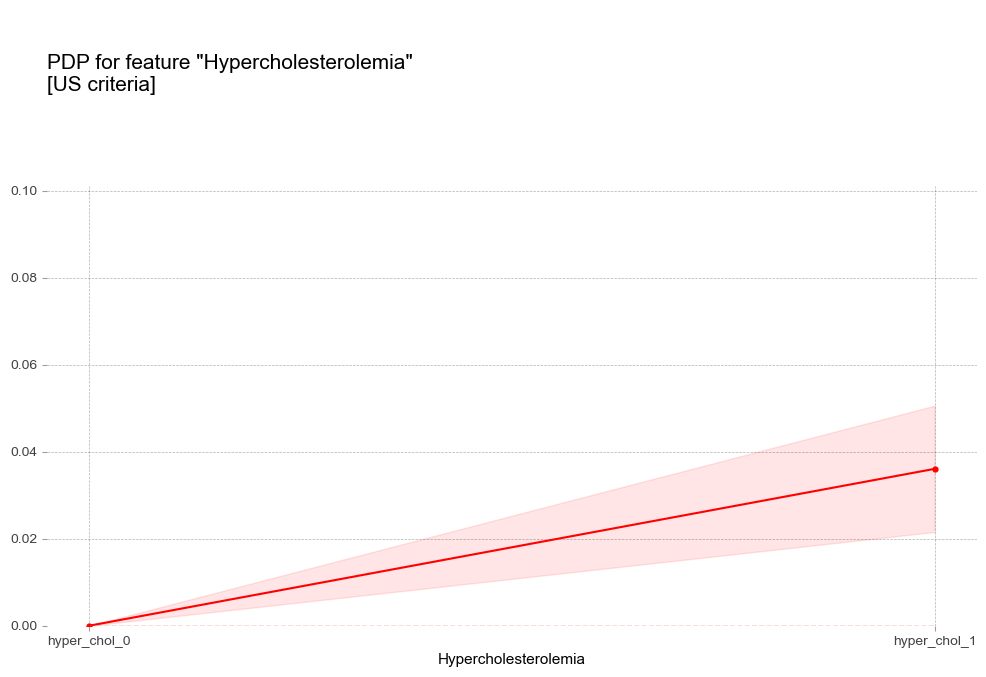
- 고콜레스테롤혈증 : 고콜레스테롤혈증이 있는 경우 더 높은 고혈압 확률을 나타냄
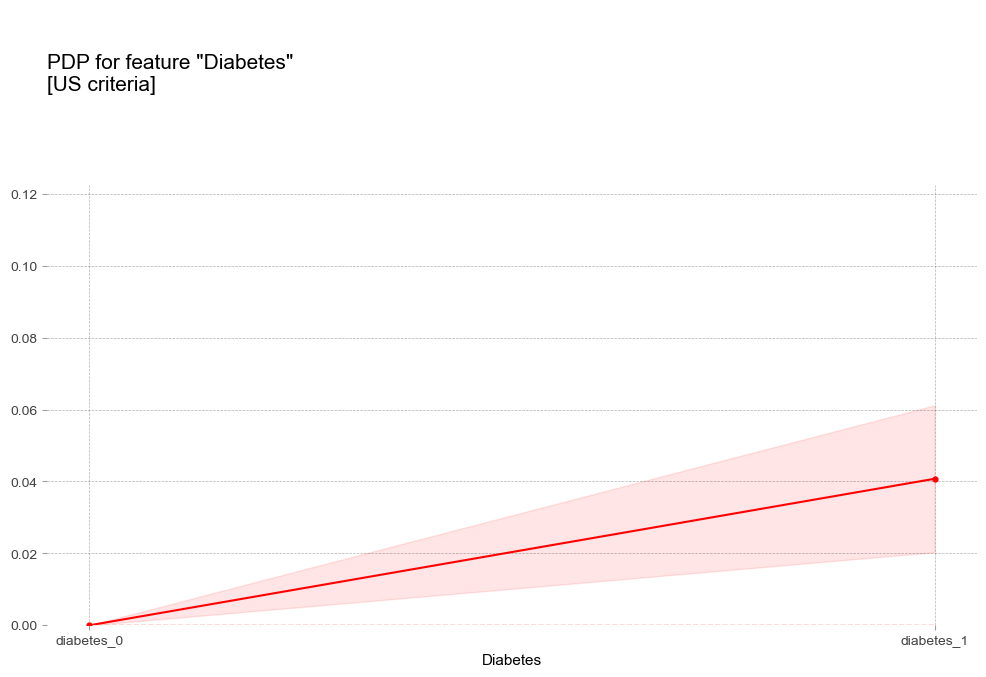
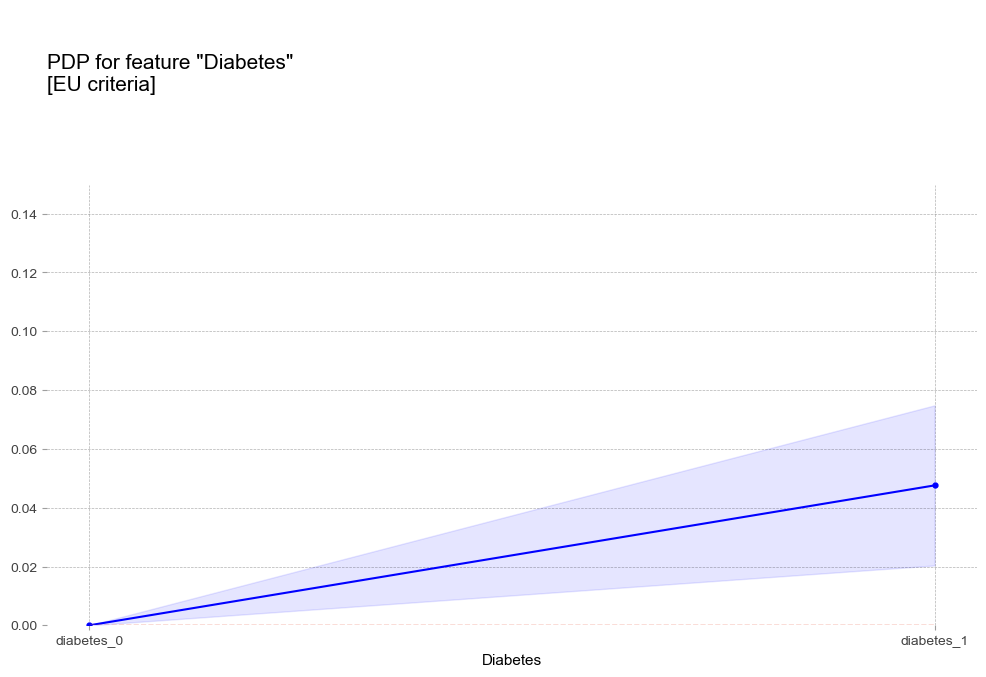
- 당뇨병 : 당뇨병이 있는 경우 더 높은 고혈압 확률을 나타냄
-
소스코드는 위에서 실시한 조절불가요인 PDP와 거의 동일하므로 그래프만 올리도록 하겠음.
-
중성지방 Triglycerides [연속형변수]
-
고콜레스테롤혈증 Hypercholesterolemia [범주형변수]
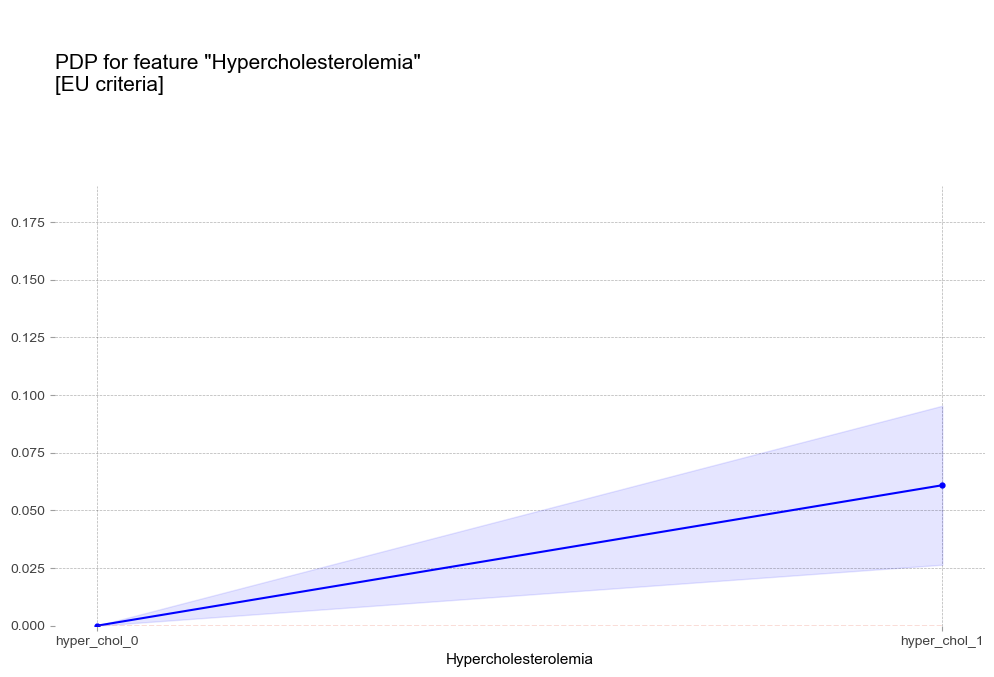
-
당뇨병 Diabetes [범주형변수]
생활습관 요인 (비만, 폭음)
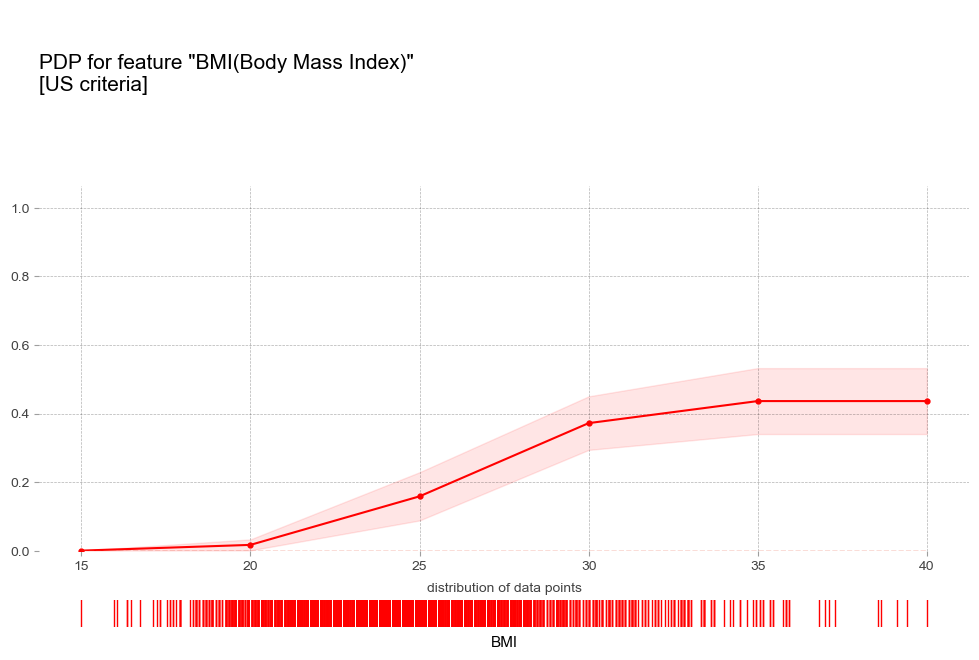
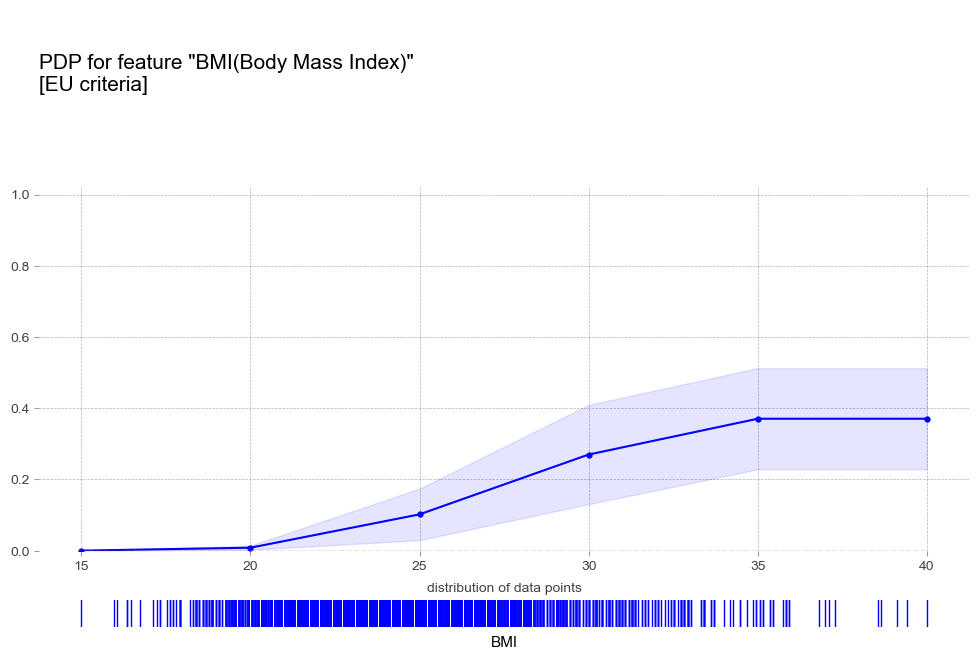
- 비만 : 비만 1단계 진단기준인 25를 기준으로 고혈압 예측 확률이 급격히 증가함
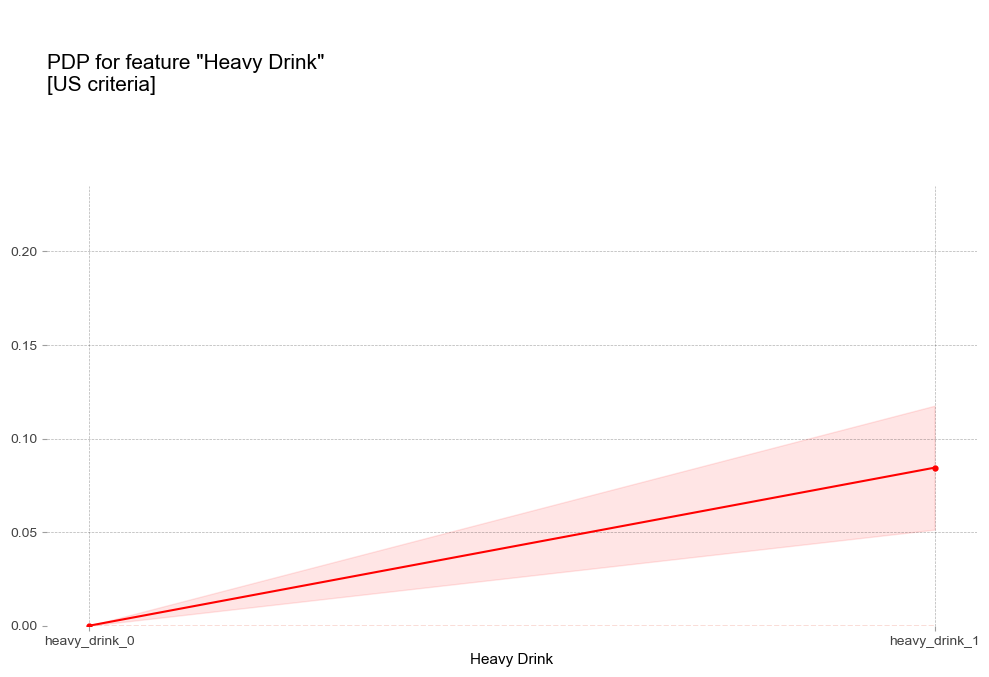
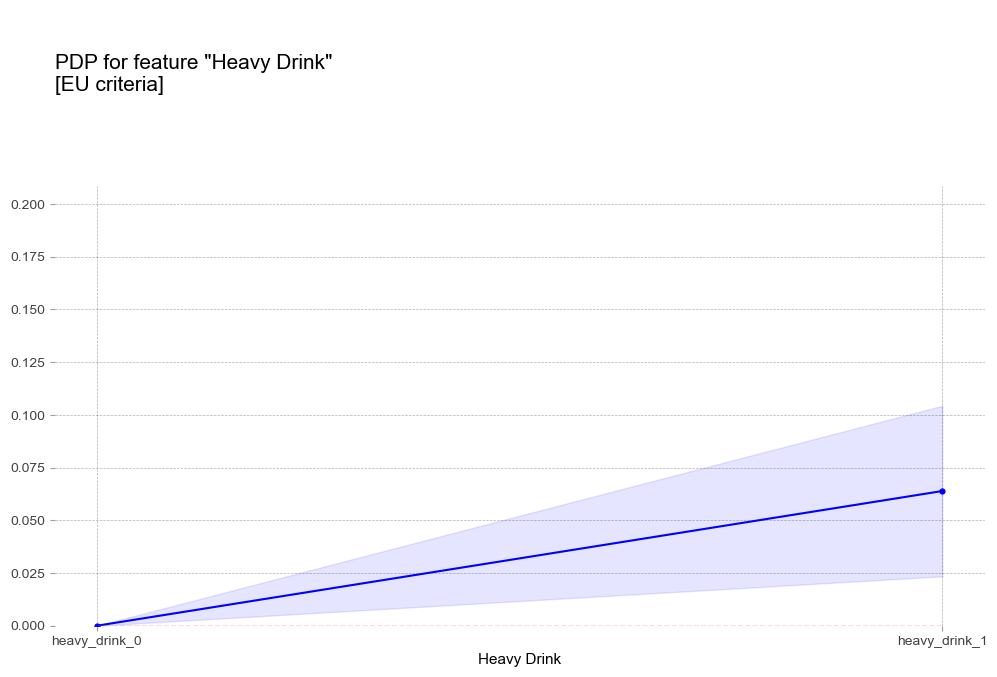
- 폭음 : 폭음을 하는 경우 더 높은 고혈압 확률을 나타냄
-
소스코드는 위에서 실시한 조절불가요인 PDP와 거의 동일하므로 그래프만 올리도록 하겠음.
-
비만도 BMI [연속형변수]
-
폭음 Heavy Drink [범주형변수]
4-4. 해석결과 요약
- 특성중요도(순열중요도)
- 미국 진단기준 (130mmHg/80mmHg)
- 나이 >> BMI >> 가족력 > 성별 >> 폭음 > 고콜레스테롤혈증 > 중성지방 >> 당뇨
- 유럽 진단기준 (140mmHg/90mmHg)
- 나이 >>> BMI >> 가족력 > 고콜레스테롤혈증 >> 폭음 > 당뇨 > 성별 > 중성지방
- 공통 중요도 TOP1 특성 : 나이
- 공통 중요도 TOP3 특성 : 나이, BMI, 가족력
- 미국 진단기준 (130mmHg/80mmHg)
- PDP(특성 경향성)
- 조절 불가 요인 (나이, 성별, 가족력)
- 나이 : 연령이 높을 수록 고혈압 예측 확률이 증가
- 성별 : 여성에비해 남성이 더 높은 고혈압 확률
- 가족력 : 가족력이 강할수록 고혈압 예측 확률이 증가
- 만성질환 요인 (중성지방, 고콜레스테롤혈증, 당뇨병)
- 중성지방 : 중성지방 수치가 높을 수록 고혈압 예측 확률이 증가
고중성지방혈증 진단기준(200mg/dL)에서 예측 확률 급증 - 고콜레스테롤혈증 : 고콜레스테롤혈증이 있는 경우 더 높은 고혈압 확률
- 당뇨병 : 당뇨병이 있는 경우 더 높은 고혈압 확률
- 중성지방 : 중성지방 수치가 높을 수록 고혈압 예측 확률이 증가
- 생활습관 요인 (비만, 폭음)
- 비만 : BMI 수치에 따라 꾸준히 예측확률 증가
비만 1단계 진단기준인 25를 기준으로 고혈압 예측 확률 급격히 증가 - 폭음 : 폭음을 하는 경우 더 높은 고혈압 확률
- 비만 : BMI 수치에 따라 꾸준히 예측확률 증가
- 조절 불가 요인 (나이, 성별, 가족력)
05. Conclusion 결론
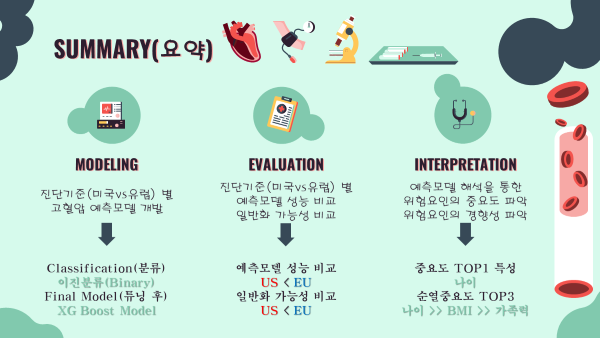
5-1. Summary 요약
- 예측모델 개발
- 이진분류문제로 정의
- 최종모델 : XGBoost (튜닝)
- 진단기준별 성능 및 일반화 가능성 비교
- 예측모델 성능 : 미국진단기준 < 유럽진단기준
- 예측모델 일반화 가능성 : 미국진단기준 < 유럽진단기준
- 예측모델 해석
- 공통 중요도 TOP1 : 나이
- 공통 순열중요도 TOP3 : 나이, 비만, 가족력
5-2. Conclusion & Limitation 결론 및 한계점
- 결론 및 인사이트
- 유럽진단기준의 모델 성능과 일반화가능성이 더 높았음
- 미국 진단기준을 따르기보단 유럽처럼 보수적인 입장을 취하는것이 더 나을 것이라 판단됨.
- 특성 중요도 TOP1 : 나이
- 고혈압 위험군 선별 및 예방 정책 수립시 고령층을 최우선으로 고려해야할 것으로 판단됨.
- 뒤이어서 BMI, 가족력이 중요한 특성
- 비만이 있거나, 고혈압 가족력이 있는 경우 고혈압 예방에 더욱 주의를 기울일 필요가 있다고 판단됨.
- 유럽진단기준의 모델 성능과 일반화가능성이 더 높았음
- 한계점
- 운동부족, 스트레스, 나트륨 과잉섭취 처럼 고혈압 유발에 중요하다 알려진 변수를 이용하지 못함
- 해당 변수에 대한 객관적 측정 및 수치화 가능한 지표가 마련되어야함.
- 복부비만 및 체중 데이터를 drop하고 분석실시함
- 비만 요인중 어떤 요인이 더 중요할지에 대한 객관적 근거가 마련되어야함.
- 가족력 변수가 기존의 유럽기준으로 기록된 변수이므로 미국진단기준과 맞지 않았을 가능성
- 미국 진단기준에 따른 가족력 판단 정보가 있었으면 더 좋았을 것이라 생각함.
- 설문조사 변수 중 모르겠다 응답한 경우를 0이라 판단하여 분석 실시함
- 조사 단계에서 좀 더 정확한 설문조사 실시가 이뤄져야 할 것이라 생각함.
- 운동부족, 스트레스, 나트륨 과잉섭취 처럼 고혈압 유발에 중요하다 알려진 변수를 이용하지 못함

5-3. Retrospective 회고

- 이번 프로젝트도 저번 프로젝트와 마찬가지로 이후 Section을 진행하느라 조금 늦게 정리가 이루어졌음...
- 이번 프로젝트에서도 준비된 데이터에 비해 정해진 발표시간이 짧아 많은 부분들을 스킵해가며 발표를 진행했었으나, 블로그 정리를 통해 전하고자 했던 내용을 모두 담아낼 수 있었음.
- 주제와 데이터셋 선정이 자유로웠던 만큼 오랜기간 생물학, 심리학, 의학 분야에서 일하고 공부해왔던 경험을 바탕으로 관심 도메인이었던 헬스케어 분야의 머신러닝 프로젝트를 진행할 수 있어서 즐거운 마음으로 프로젝트에 임할 수 있었던 점이 너무 좋았음.
- 물론 프로젝트를 구상하고 데이터를 선정하는데 다소 스트레스를 받긴 했으나, 기획 및 데이터 선정을 프로젝트 실시 기간 2~3주 전부터 진행해와서 비교적 완성도 있는 결과물을 낸것 같아 뿌듯했음.
- 프로젝트 내용을 정리하면서 배웠던 내용들을 복습할 수 있는 유의미한 시간이었고, 코드 구성도 전반적으로 만족스러웠다 생각함.
- 코치진에게 받은 한가지 지적사항은 너무 많은 내용을 발표에 담으려 해서 EDA나 모델링에 대한 설명이 부족했던 점이었음. 스킵해서 발표를 진행했으나 발표 자료에 해당 내용들이 잘 담겨져 있어서 문제가 되진 않았다하여, 프로젝트 자체의 문제점은 딱히 없는 듯함. 고생한만큼 완성도가 높게 평가되어 정말 뿌듯했음.
- 본 프로젝트 정리 글을 쓴 시점이 딥러닝을 배우기 직전에 쓴 것이라 그동안 배운것을 되돌아보고 정리한 것이 복습하는데 정말 도움이 많이 된 것 같고, 앞으로도 진행한 프로젝트에 대해선 정리해보는 시간을 가져야겠다는 생각이 들었음. 정리하는데 시간이 생각보다 많이 걸리긴 했지만 의미는 충분히 있었다고 생각함.
- 아직도 배울게 산더미지만... 포기하지 않고 교육과정을 잘 마무리할 수 있었으면 좋겠고, 더욱 열심히 해야겠다는 다짐을 다시한번 해볼 수 있는 시간이었음.
- 아자아자 파이팅!
