3. 결정트리 분류모델 활용한 유방암 예측
0) 문제정의
- DecisionTreenClassifier 사용하여 유방암 양성(2), 악성(4) 예측
1) 라이브러리 임포트
from sklearn import preprocessing # 정규화 포함한 전처리 관련
from sklearn.model_selection import train_test_split # 데이터셋 분리 관련
from sklearn import tree # 결정트리 모델 관련
from sklearn import metrics # 성능평가 관련
import pandas as pd
import numpy as np2) 데이터 준비하기
uci_path = 'https://archive.ics.uci.edu/ml/machine-learning-databases/\
breast-cancer-wisconsin/breast-cancer-wisconsin.data'
df = pd.read_csv(uci_path, header = None)
df.head()- url을 변수에 담고 변수로 데이터 셋을 읽어 들인다.
- 'header = None'은 컬럼명이 없다는 뜻이고 판다스는 0부터 순서대로 header를 부여한다.
- 데이터셋 출처: https://archive.ics.uci.edu/ml/machine-learning-databases//breast-cancer-wisconsin/

# 열 이름 지정하기
df.columns = ['id', 'clump', 'cell_size', 'cell_shape', 'adhesion', 'epithlial',
'bare_nuclei', 'chromatin', 'normal_nucleoli', 'mitoses', 'class']
df.head()- 데이터셋 출처 웹사이트에서 컬럼명을 확인해 열 이름을 지정해준다.

3) 데이터 확인하기
-df.info()로 확인해보니 총 699건의 데이터가 있고 다른 컬럼은 정수형(숫자형)이었지만, 'bare_nuclei'라는 컬럼만 object type이었다. 머신러닝 모델은 숫자만 이해하고 처리하기 때문에 전처리가 필요하다.
# 1) 'bare_nuclei'의 고유값(unique) 확인
df['bare_nuclei'].unique()- 결과값: array(['1', '10', '2', '4', '3', '9', '7', '?', '5', '8', '6'], dtype=object)
- '?' 때문에 object 타입이 되었다는 것을 알 수 있다.
# 2) '?' -> np.nana으로 변경하고 수를 확인하자
df['bare_nuclei'].replace('?', np.nan, inplace = True)
df['bare_nuclei'].isna().sum()- 결과값은 16으로 총 NaN값이 16개임을 알 수 있다.
# 3) NaN값을 삭제하고 전체 수 확인
df.dropna(subset=['bare_nuclei'], axis =0, inplace = True)
df.info()- 해당 열에서 NaN값이 있다면 그 값이 속한 행을 모두 지워준 후 데이터 수를 확인해보니 683개로 16개의 NaN값이 모두 지워진 것을 확인함
# 4) bare_nuclei 컬럼의 형변환
df['bare_nuclei'] = df['bare_nuclei'].astype('int64')
df.info()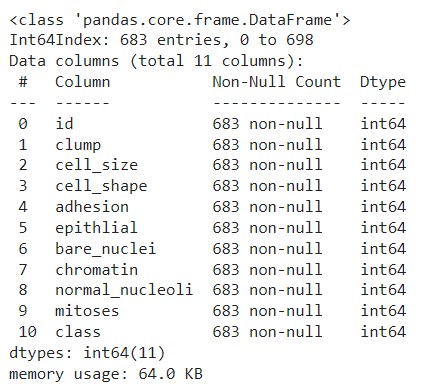
- 모든 컬럼의 데이터 타입이 정수형이 되었음
4) 데이터 분리하기
X = df[['clump', 'cell_size', 'cell_shape', 'adhesion', 'epithlial',
'bare_nuclei', 'chromatin', 'normal_nucleoli', 'mitoses']]
y = df['class']- id는 유방암의 특징이 아닌 unique한 값이기 때문에 X feature에서 제외
X
- X를 출력해보면 각 데이터들의 구간이 넓기 때문에 머신러닝이 학습하기 어려운 조건이다. 따라서 정규화->스케일링이 필요하다.
#스케일링 후 데이터 분리
X = preprocessing.StandardScaler().fit(X).transform(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state = 7)
print('train shape', X_train.shape)
print('test shape', X_test.shape)- 결과값:
train shape (478, 9)
test shape (205, 9)
5) 결정트리 분류모델 설정
tree_model = tree.DecisionTreeClassifier(criterion= 'entropy', max_depth=5)- 모델 객체 생성(최적의 속성을 찾기 위해 criterion='entropy'를 적용하여 계산) -> 적정한 레벨 값 찾는 것이 중요
- 최대 가지 깊이를 5개로 규제함 -> 강력한 사전 가지치기
6) 모델 학습하기 및 예측하기
tree_model.fit(X_train, y_train)
y_pred = tree_model.predict(X_test)7) 모델 성능평가
print('훈련 세트 정확도: {:.2f}%'.format(tree_model.score(X_train, y_train)*100))
print('테스트 세트 정확도: {:.2f}%'.format(tree_model.score(X_test, y_test)*100))- 소수점 둘째자리까지 정확도를 측정
- 결과값:
훈련 세트 정확도: 98.33%
테스트 세트 정확도: 94.63%
8) 모델 성능평가 지표 계산
tree_report = metrics.classification_report(y_test, y_pred)
print(tree_report)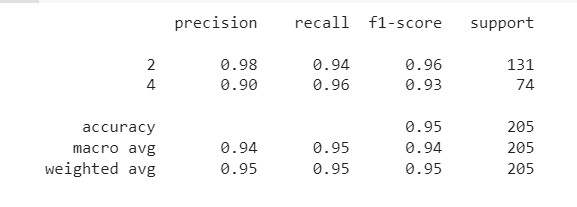
- 악성의 정확도는 90%, 재현율은 96%임을 알 수 있다.
9) 결정 트리 그래프 그리기
from sklearn.tree import export_graphviz # 트리 모델의 그래프
export_graphviz(tree_model, out_file='tree.dot', class_names=['악성','양성'],
feature_names=df.columns[1:10], impurity=False, filled=True)
import graphviz
with open('tree.dot') as f:
dot_graph = f.read()
display(graphviz.Source(dot_graph)) 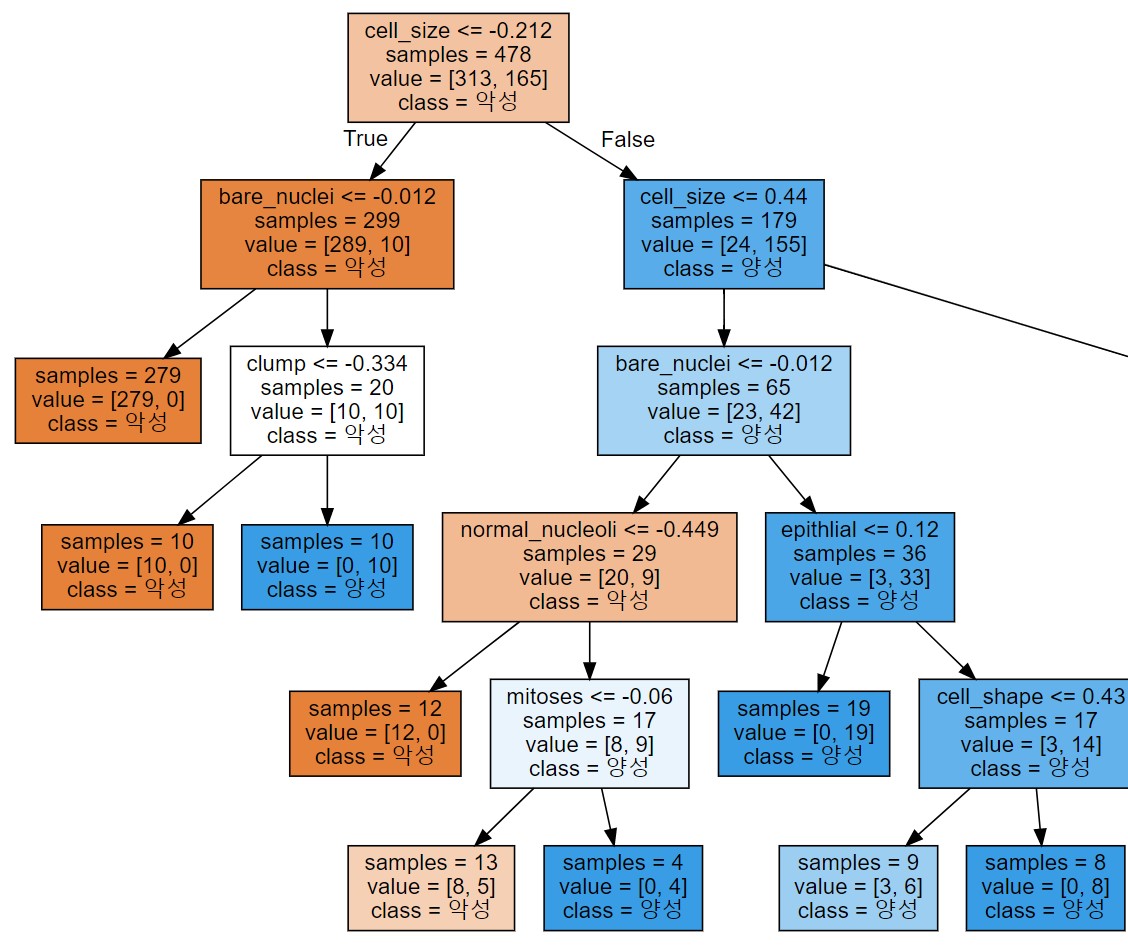
- 가지가 2개 적어도 3개만 그려져도 대다수의 악성을 분류해낼 수 있다는 것을 알 수 있다.
- 답정너 같은 스타일이라 학습 데이터에 과대적합되는 경향이 강하다.(결정트리 단점)
10) 그래프 그려서 최적의 가지수 찾기
train_scores = []
test_scores = []
max_depth = np.arange(1, 10, 1) #가지 개수 1-10까지 설정
for n in max_depth:
# 모델 생성 및 학습
tree_model = tree.DecisionTreeClassifier(criterion= 'entropy', max_depth=n).fit(X_train, y_train)
# 훈련 세트 정확도 저장
train_scores.append(tree_model.score(X_train, y_train))
# 테스트 세트 정확도 저장
test_scores.append(tree_model.score(X_test, y_test))
# 예측 정확도 비교 그래프 그리기
plt.figure(dpi=150)
plt.plot(max_depth, train_scores, label='훈련 정확도')
plt.plot(max_depth, test_scores, label='테스트 정확도')
plt.ylabel('정확도')
plt.xlabel('가지개수')
plt.legend()
plt.show() 
- 가지개수는 3이 적정하다는 것을 확인할 수 있다.
4. 앙상블 분류모델 활용한 심장병 예측
0) 문제 정의
- 환자 정보를 사용하여 심장 질환 유무(정상:0, 심장질환 진단:1)를 예측하는 이진 분류 문제로 정의
- 데이터 출처: https://archive.ics.uci.edu/ml/datasets/heart+disease
1) 라이브러리 임포트
import numpy as np # 데이터 처리 관련
import pandas as pd
import matplotlib.pyplot as plt # 시각화 관련
import plotly.express as px
import seaborn as sns
from sklearn.model_selection import train_test_split # 데이터 셋 분리 관련
from sklearn.preprocessing import StandardScaler # 데이터 정규화 관련
from sklearn.linear_model import LogisticRegression # 로지스틱 회귀 모델
from sklearn.tree import DecisionTreeClassifier # 결정트리 분류 모델
from sklearn.ensemble import RandomForestClassifier # 랜덤포레스트 분류 모델(앙상블)
from sklearn.ensemble import GradientBoostingClassifier # 그래디언트부스팅 분류 모델(앙상블)
from sklearn import metrics # 성능평가df = pd.read_csv('/content/heart.csv')
df.head()
- csv 파일을 읽어들이고 데이터 확인해보니 두가지 전처리가 필요해보임
- 구간이 넓은 숫자형은 정규화 처리, 범주형은 인코딩
2) 데이터 전처리
df['sex'].unique()- 0아니면 1인 것을 보아 범주형임 -> 원핫인코딩하자
df['cp'].unique() - 0 - 3 통증의 종류 -> 원핫인코딩 하자
df['fbs'].unique() - 0아니면 1인 것을 보아 범주형임 -> 원핫인코딩하자
categorical_var = ['sex', 'cp', 'fbs', 'restecg', 'exng', 'slp', 'caa', 'thall']
df[categorical_var] = df[categorical_var].astype('category') - 1) 카테고리(범주형) 컬럼 -> Dtpe 변경 -> 원핫인코딩 -> 예시. sex, fbs 컬럼
- 범주형 데이터 컬럼을 카테고리형 변수에 담고, 숫자형 -> 카테고리 타입으로 변경
numberic_var = [i for i in df.columns if i not in categorical_var][:-1]- 2) 숫자형(연속형) 컬럼 -> 정규화 필요
- df의 컬럼 중 카테고리형 변수에 포함되지 않은 컬럼은 숫자형 변수에 담기
X= df.iloc[:, :-1]# 앞: data row 수 뒤: column 수(':'=전부를 의미)
y = df['output']- 인코딩과 정규화를 한 번에 처리하기 위해 문제지와 정답지에 먼저 데이터를 담는다.
- df의 마지막 열이자 정답지인 output을 X feature값에서 제외한다.
#범주형 데이터 전처리
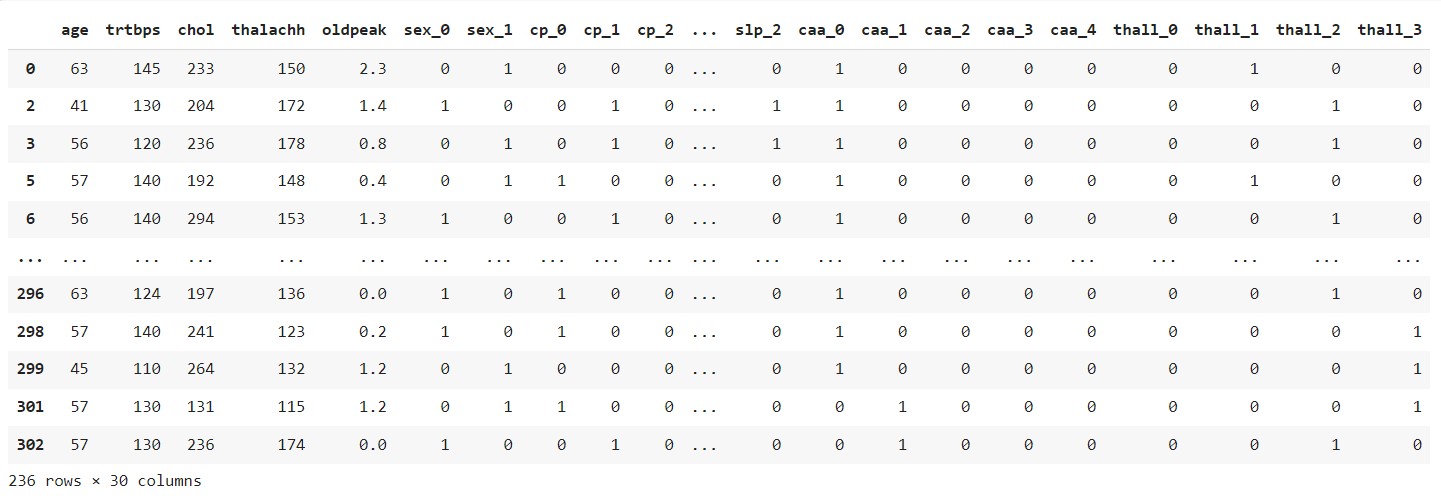
temp_X = pd.get_dummies(X[categorical_var])
X_modified = pd.concat([X, temp_X], axis =1)
X_modified.drop(categorical_var, axis=1, inplace =True)
X_modified- 1) 범주형 컬럼들 -> 원핫인코딩
- 2) 원핫인코딩 컬럼 추가하여 기존 df와 열 기준 머지
- 3) 기존 범주형 컬럼 삭제
#수치형 변수 전처리
X_modified[numberic_var] = StandardScaler().fit(X_modified[numberic_var]).transform(X_modified[numberic_var])- 구간이 넓었던 데이터를 0과 1 사이의 값으로 스케일링
3) 데이터 분리하기
X_train, X_test, y_train, y_test = train_test_split(X_modified, y, test_size=0.2, random_state = 7)
print('train shape', X_train.shape)
print('test shape', X_test.shape)- 결과값:
train shape (188, 30)
test shape (48, 30)
4) 머신러닝 모델 설정 및 학습
# 1. LogisticRegression
logreg = LogisticRegression().fit(X_train, y_train)
print('훈련 세트 정확도: {:.5f}%'.format(logreg.score(X_train, y_train)*100))
print('테스트 세트 정확도: {:.5f}%'.format(logreg.score(X_test, y_test)*100))- 결과값:
훈련 세트 정확도: 90.42553%
테스트 세트 정확도: 83.33333%
# 2. DecisionTree
tree = DecisionTreeClassifier(max_depth=5, min_samples_leaf=20, min_samples_split=40).fit(X_train, y_train)
print('훈련 세트 정확도: {:.5f}%'.format(tree.score(X_train, y_train)*100))
print('테스트 세트 정확도: {:.5f}%'.format(tree.score(X_test, y_test)*100))- 결과값:
훈련 세트 정확도: 81.91489%
테스트 세트 정확도: 81.25000%
# 3. RandomForest
# 결정트리와 달리 랜덤하게 가지치기 시작 feature를 선택한다.
random = RandomForestClassifier(n_estimators=300, random_state=7).fit(X_train, y_train)
print('훈련 세트 정확도: {:.8f}%'.format(random.score(X_train, y_train)*100))
print('테스트 세트 정확도: {:.8f}%'.format(random.score(X_test, y_test)*100))- 결과값:
훈련 세트 정확도: 100.00000000%
테스트 세트 정확도: 85.41666667% - 현재 훈련 정확도가 100%라는 건 과대적합이라는 뜻
- 조정 가능 값: n_estimators
# 4. GradientBoosting
boost = GradientBoostingClassifier(max_depth=3, learning_rate=0.05).fit(X_train, y_train)
print('훈련 세트 정확도: {:.8f}%'.format(boost.score(X_train, y_train)*100))
# 패턴 일반화로 훈련 데이터에 대한 정확도는 떨어짐
print('테스트 세트 정확도: {:.8f}%'.format(boost.score(X_test, y_test)*100))- 결과값:
훈련 세트 정확도: 98.40425532%
테스트 세트 정확도: 87.50000000% - 현재 훈련 정확도를 보아 과대적합이므로 강력한 사전 가지치기 필요
- 조정 가능 값: max_depth, learning_rate
