Timeseries Anomaly Detection-(3) Unsupervised Learning
UnSupervised Learning
1. z-test
Check the distribution of datasets
# Check the distribution of the above stock data
fig, ax = plt.subplots(figsize=(9,6))
_ = plt.hist(df.Close, 100, density=True, alpha=0.75)- Result
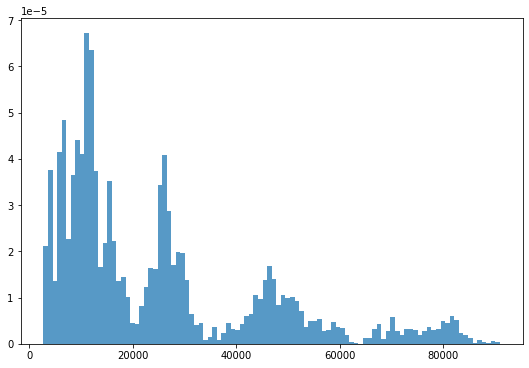
It does not follow a normal distribution.
from statsmodels.stats.weightstats import ztest
_, p = ztest(df.Close)
print(p) # 0If the value of p is less than 0.05, it means that it is far from the normal distribution, so it is difficult to analyze the confidence interval assuming the normal distribution with this data.
2) To extract data close to a normal distribution from a time series data set
from statsmodels.tsa.seasonal import seasonal_decompose
# Assume that the term of seasonality is 50 days
# extrapolate_trend='freq' : Because of the rolling window for creating a trend, the Nan value inevitably occurs in trend, resid. Therefore, it is an option that fills this NaN value.
result = seasonal_decompose(df.Close, model='additive', two_sided=True,
period=50, extrapolate_trend='freq')
result.plot()
plt.show()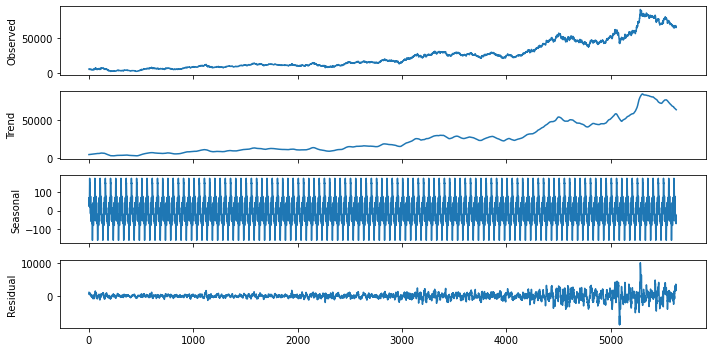
Check the 3rd Seasonal component in detail
result.seasonal[:100].plot()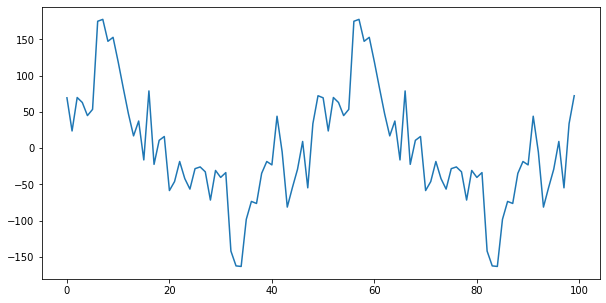
→ It can be seen that it repeats periodically between -160 and 180.
Since residuals are distributed based on an average of 0, days with large residuals can be interpreted as days out of general trend or seasonality.
# Check the distribution of Residual
fig, ax = plt.subplots(figsize=(9,6))
_ = plt.hist(result.resid, 100, density=True, alpha=0.75)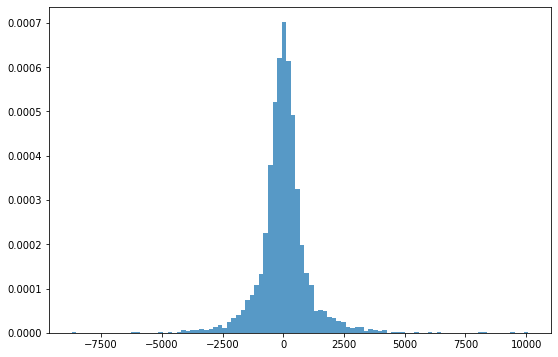
z-test (Check by number)
r = result.resid.values
st, p = ztest(r)
print(st,p)Result
0.6948735022171139 0.4871345798943748Since the value of p is more than 0.05 and the graph follows a normal distribution, the confidence interval analysis can be performed using this dataset.
# Output of Average and Standard Deviation
mu, std = result.resid.mean(), result.resid.std()
print("평균:", mu)
print("표준편차:", std)
# Determine outliers based on 3-sigma (standard deviation)
print("이상치 갯수:", len(result.resid[(result.resid>mu+3*std)|(result.resid<mu-3*std)]))
#result:
#mean: 9.368886923797055
#standard deviation: 1011.6643445235127
#Number of outliers: 112To view the details of 112 outliers
df.Date[result.resid[(result.resid>mu+3*std)|(result.resid<mu-3*std)].index]4385 2017-05-08
4436 2017-07-20
4452 2017-08-11
4453 2017-08-14
4467 2017-09-04
...
5619 2022-05-20
5620 2022-05-23
5625 2022-05-30
5626 2022-05-31
5628 2022-06-03
Name: Date, Length: 112, dtype: datetime64[ns]Due to the explosion of Galaxy Note 7 in 2017 and the recent plunge in stock prices, abnormalities can be found.
# Data preprocessing
def my_decompose(df, features, freq=50):
trend = pd.DataFrame()
seasonal = pd.DataFrame()
resid = pd.DataFrame()
# Decompose for each feature to be used.
for f in features:
result = seasonal_decompose(df[f], model='additive', freq=freq, extrapolate_trend=freq)
trend[f] = result.trend.values
seasonal[f] = result.seasonal.values
resid[f] = result.resid.values
return trend, seasonal, resid
# Trends/seasonal/residuals for each variable
tdf, sdf, rdf = my_decompose(df, features=['Open','High','Low','Close','Volume'])
tdf.describe()result
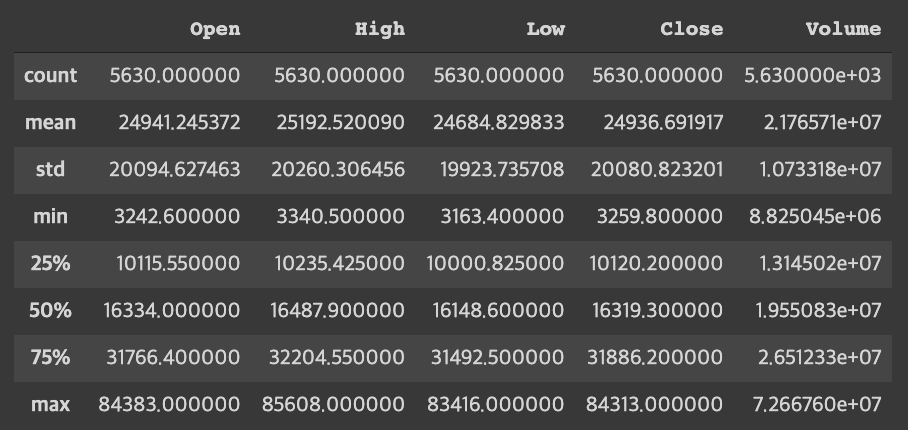
Trend by each variable
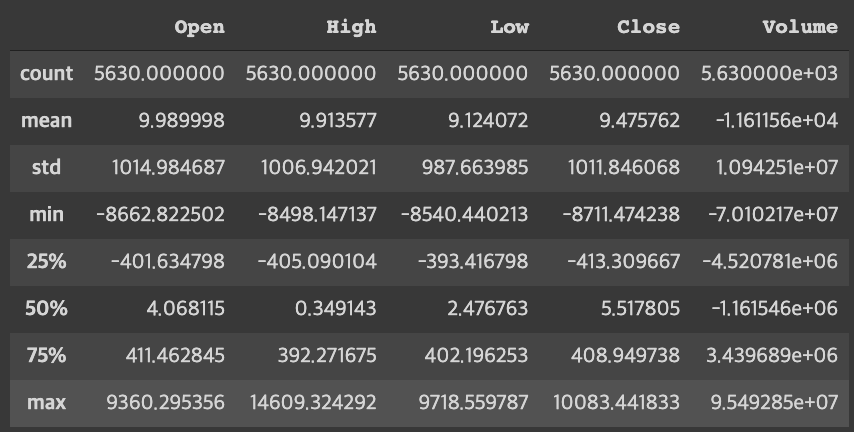
- Looking at the residuals for each variable, it can be seen that the value of the volume is particularly large compared to other values. If we use this data as it is, the volume will be reflected most importantly, so we can't get the exact value we want, so we give standard normalization.
# Standard normalization
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(rdf)
print(scaler.mean_)
norm_rdf = scaler.transform(rdf)
norm_rdfresult:
[ 9.98999848e+00 9.91357726e+00 9.12407232e+00 9.47576165e+00 -1.16115582e+04]
array([[ 0.91049378, 0.94485457, 0.76319233, 1.06571492, 2.06343412],
[ 0.73231584, 0.91140349, 0.62369328, 0.56867739, 1.93682018],
[ 0.66407391, 0.5997526 , 0.64622584, 0.54419678, -0.03804234],
...,
[ 2.80963702, 2.6765217 , 2.93000737, 2.82886965, -0.09875707],
[ 3.59222865, 3.16722954, 3.50891551, 3.09579198, -0.82225883],
[ 2.75976516, 2.3669442 , 2.24740122, 1.90013435, 0.28431761]])