캐글 스터디를 진행하면서 매 에피소드마다 얻은 인사이트나 팁 등을 정리해야지!
...라고 생각했었는데 쉽지않았고..ㅠ

벌써 ep13이 되어버렸다😅
이쯤에서 팀원들 모두 정리하고 가는 타임을 가지기로해서 이번 기회에 정리해두기.
기억에 남거나, 나중에 또 활용해볼 부분 등등 위주로 정리하기.
참여한 에피소드 돌아보니까 매번 시각화를 진짜 너무나도 열심히 봤구나 싶다.
정리해두는데 파일 이름에 EDA가 빠지지 않고 들어가는듯..
Kaggle Series3 Episode 03
Binary Classification with a Tabular Employee Attrition Dataset
plotly 사용해서 시각화하기
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.figure_factory as ffBar chart (Missing Value Visualization)
# Attrition = Target
data = pd.DataFrame(len(train.Attrition) - train.isnull().sum(), columns=['Count']
# Bar chart로 시각화
trace = go.Bar(x=data.index, y=data['Count'], # Bar chart
opacity=0.8,
marker=dict(color='lightgrey', # Bar 색, 선, 두께 지정
line=dict(color='#444444'), width=1.5))
layout = dict(title='Missing Values')
fig = dict(data=[trace], layout=layout)
py.iplot(fig)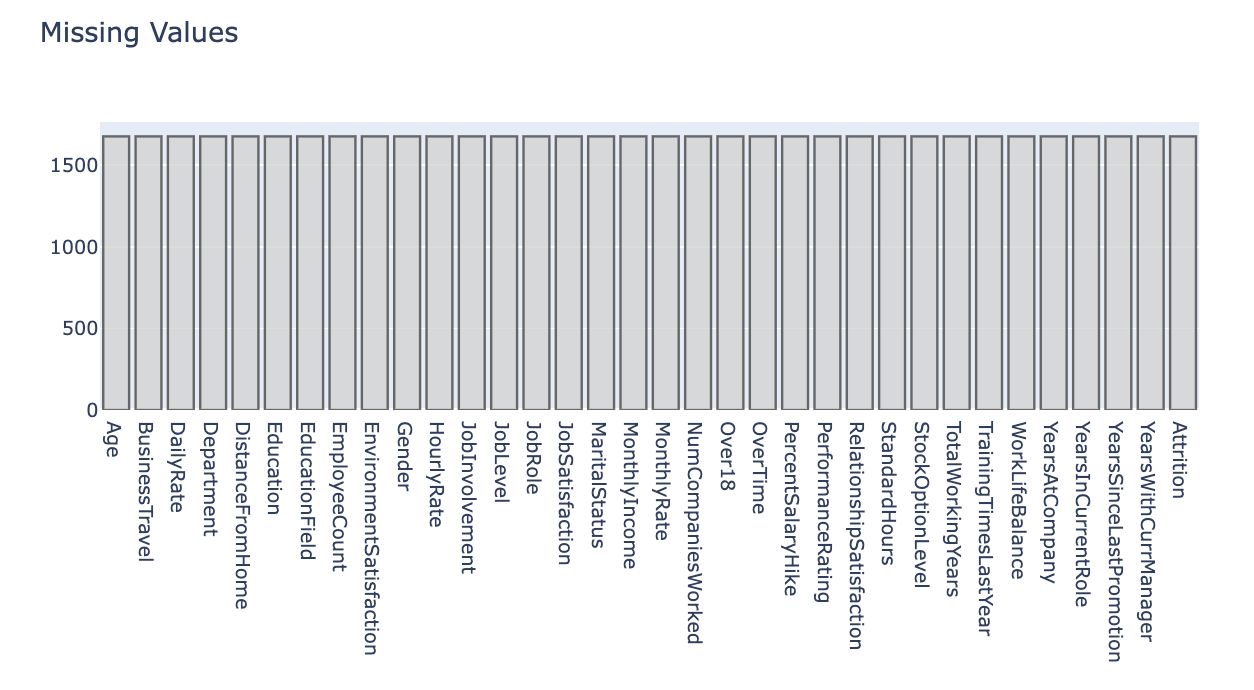
Pie Chart (Target Distribution)
# target 값의 분포 확인
target = train[train.Target != 0]
no_target = train[train.Target == 0]
# Pie Chart로 PERCENT 표현
trace = go.Pie(labels=['no_Target', 'Target'],
values=train.Target.value_counts(),
textfont=dict(size=15), opacity=0.8,
marker=dict(color=['lightskyblue','gold'],
line=dict(color='#000000',width=1.5)))
layout = dict(title='Distribution of Target variabble')
fig = dict(data=trace, layout=layout)
py.iplot(fig)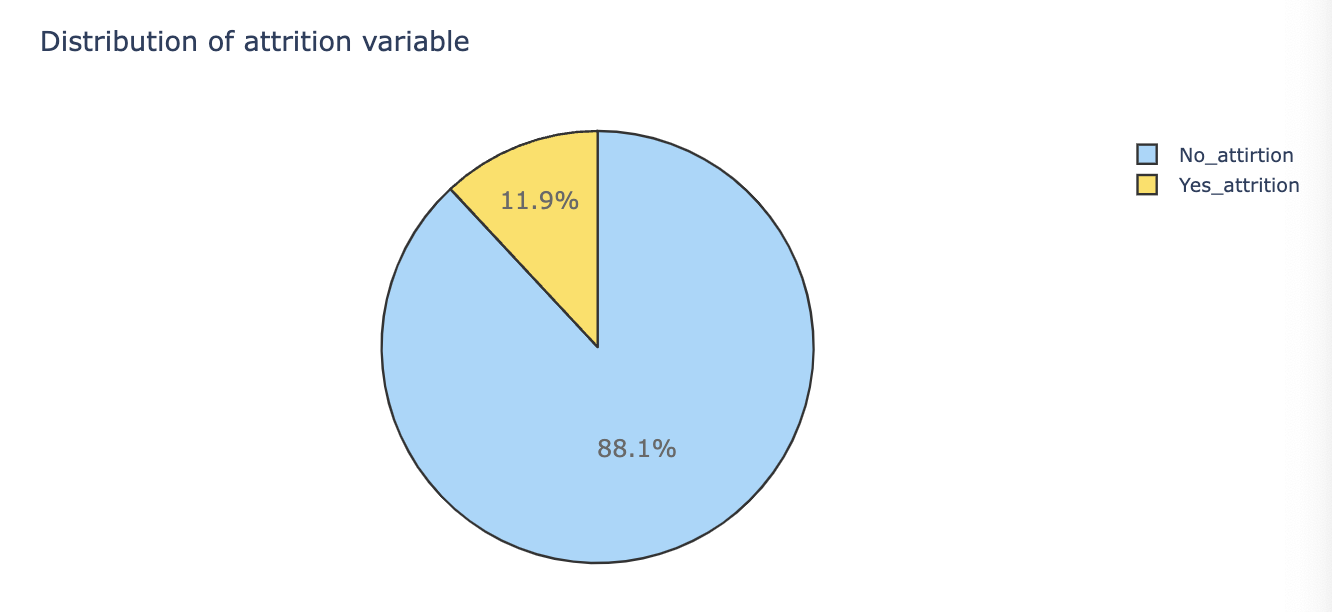
Distplot (Features Distribution)
corr = train.Target.corr(train['Features'])
corr = np.round(corr,3)
tmp1 = target['Features']
tmp2 = no_target['Features]
hist_data = [tmp1, tmp2]
group_labels = ['Target', 'no_Target']
colors = ['#FFD700', '#7EC0EE']
fig = ff.create_distplot(hist_data, group_labels, colors=colors, show_hist=True, curve_type='kde', bin_size=False)
fig['layout'].update(title=f'Features (corr target = {corr})')
py.iplot(fig)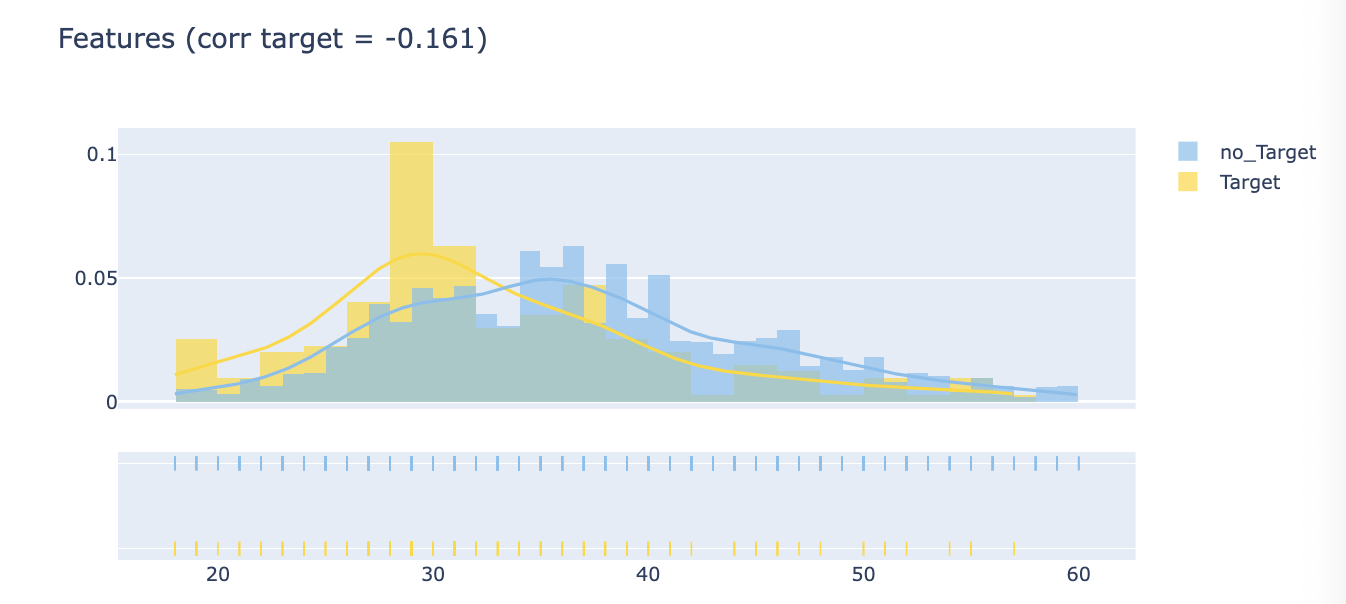
Scatter plot (mode=lines)
tmp3 = pd.DataFrame(pd.crosstab(train['Features'], train.Target), )
tmp3['target%'] = tmp3[1] / (tmp3[0] + tmp3[1]) * 100
trace3 = go.Scatter(
x=tmp3.index,
y=tmp3['target%'],
mode='lines',
name='% Target', opacity=0.6, marker=dict(color='black')
)
layout = dict(title='Features scatter lines mode')
fig = dict(data=trace3, layout=layout)
py.iplot(fig)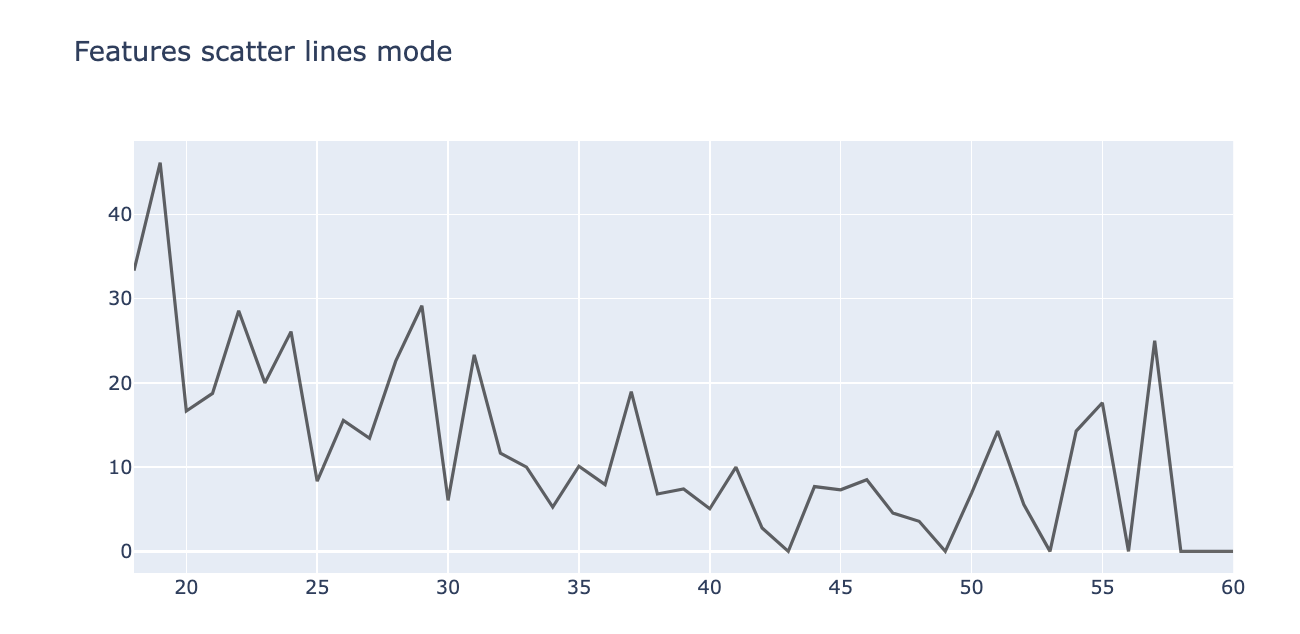
kaggle 노트북에서 참고한 plot 함수들.
def plot_distribution(var_select, bin_size):
corr = train_data.Attrition.corr(train_data[var_select])
corr = np.round(corr, 3)
tmp1 = attrition[var_select]
tmp2 = no_attrition[var_select]
hist_data = [tmp1, tmp2]
group_labels = ['Yes_attrition', 'No_attrition']
colors = ['#FFD700', '#7EC0EE']
fig = ff.create_distplot(hist_data, group_labels, colors=colors, show_hist=True, curve_type='kde', bin_size=bin_size)
fig['layout'].update(title=var_select+' '+'(corr target ='+str(corr)+')')
py.iplot(fig, filename='Density plot')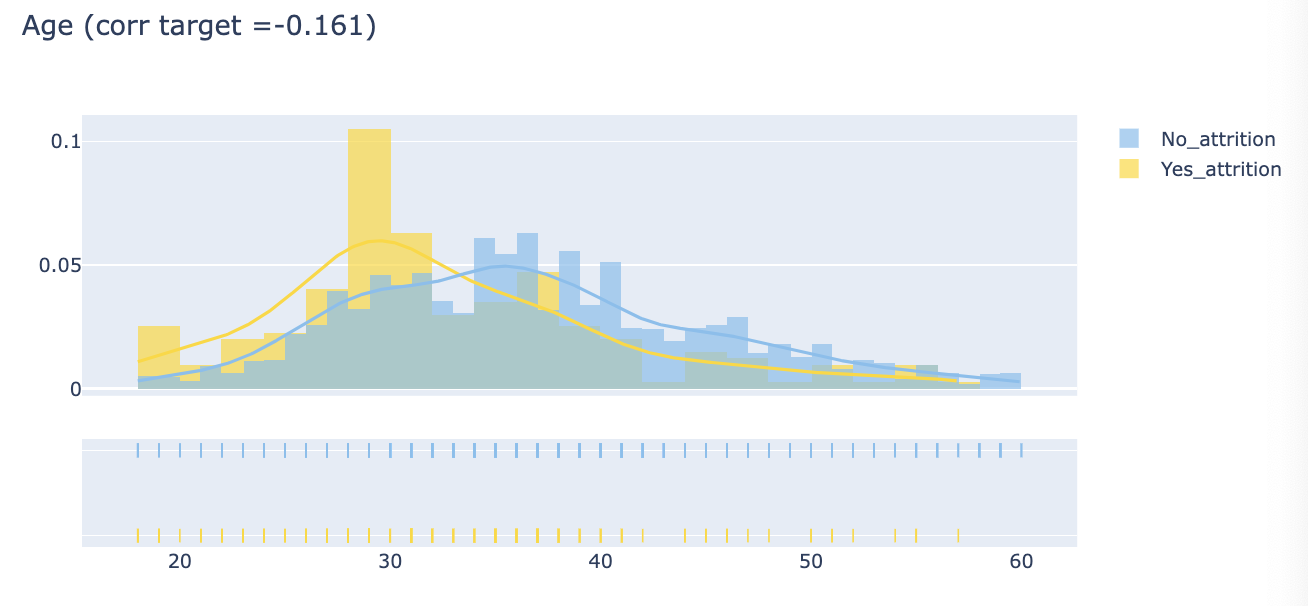
def barplot(var_select, x_no_numeric):
tmp1 = train_data[train_data.Attrition != 0]
tmp2 = train_data[train_data.Attrition == 0]
tmp3 = pd.DataFrame(pd.crosstab(train_data[var_select], train_data.Attrition), )
tmp3['Attr%'] = tmp3[1] / (tmp3[1] + tmp3[0]) * 100
if x_no_numeric == True:
tmp3 = tmp3.sort_values(1, ascending=False)
color=['lightskyblue', 'gold']
trace1 = go.Bar(
x=tmp1[var_select].value_counts().keys().tolist(),
y=tmp1[var_select].value_counts().values.tolist(),
name='Yes_Attrition', opacity=0.8, marker=dict(color='gold', line=dict(color='#000000', width=1))
)
trace2 = go.Bar(
x=tmp2[var_select].value_counts().keys().tolist(),
y=tmp2[var_select].value_counts().values.tolist(),
name='No_Attrition', opacity=0.8, marker=dict(
color='lightskyblue',
line=dict(color='#000000', width=1))
)
trace3 = go.Scatter(
x=tmp3.index,
y=tmp3['Attr%'],
yaxis = 'y2',
name='% Attrition', opacity=0.6, marker=dict(color='black', line=dict(color='#000000', width=0.5))
)
layout = dict(title = str(var_select),
xaxis=dict(),
yaxis=dict(title='Count'),
yaxis2=dict(range=[-0, 75],
overlaying = 'y',
anchor = 'x',
side = 'right',
zeroline = False,
showgrid = False,
title = '% Attrition'
))
fig = go.Figure(data=[trace1, trace2, trace3], layout=layout)
py.iplot(fig)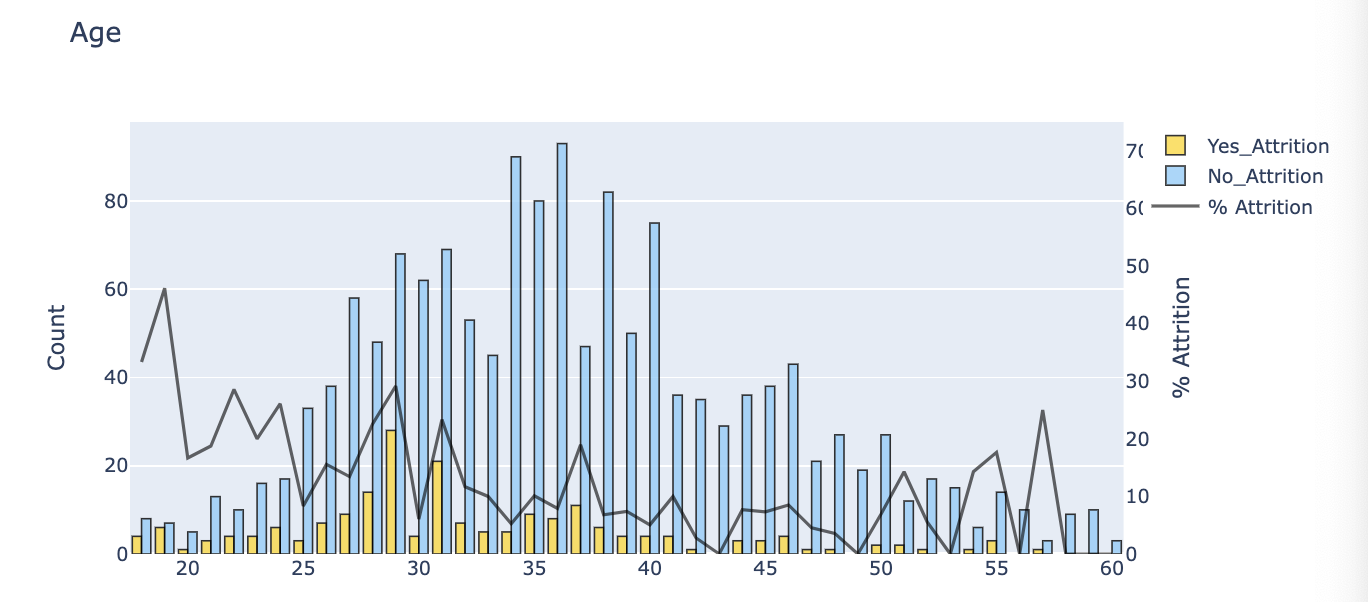
def plot_pie(var_select):
colors = ['gold', 'lightgreen', 'lightcoral', 'lightskyblue', 'lightgrey', 'orange', 'white', 'lightpink']
trace1 = go.Pie(values = attrition[var_select].value_counts().values.tolist(),
labels = attrition[var_select].value_counts().keys().tolist(),
textfont = dict(size=15), opacity = 0.8,
hoverinfo = 'label+percent+name',
domain = dict(x = [0, .48]),
name = 'attrition emplyes',
marker = dict(colors = colors, line = dict(width = 1.5)))
trace2 = go.Pie(values = no_attrition[var_select].value_counts().values.tolist(),
labels = no_attrition[var_select].value_counts().keys().tolist(),
textfont = dict(size=15), opacity = 0.8,
hoverinfo = 'label+percent+name',
marker = dict(colors = colors, line = dict(width = 1.5)),
domain = dict(x = [.52, 1]),
name = 'Non attrition employes')
layout = go.Layout(dict(title = var_select + ' distribution in employes attrition ',
annotations = [dict(text = 'Yes_attrition',
font = dict(size = 13),
showarrow = False,
x = .22, y = -0.1),
dict(text = 'No_attrition',
font = dict(size = 13),
showarrow = False,
x=.8, y=-.1)]
))
fig = go.Figure(data = [trace1, trace2], layout=layout)
py.iplot(fig)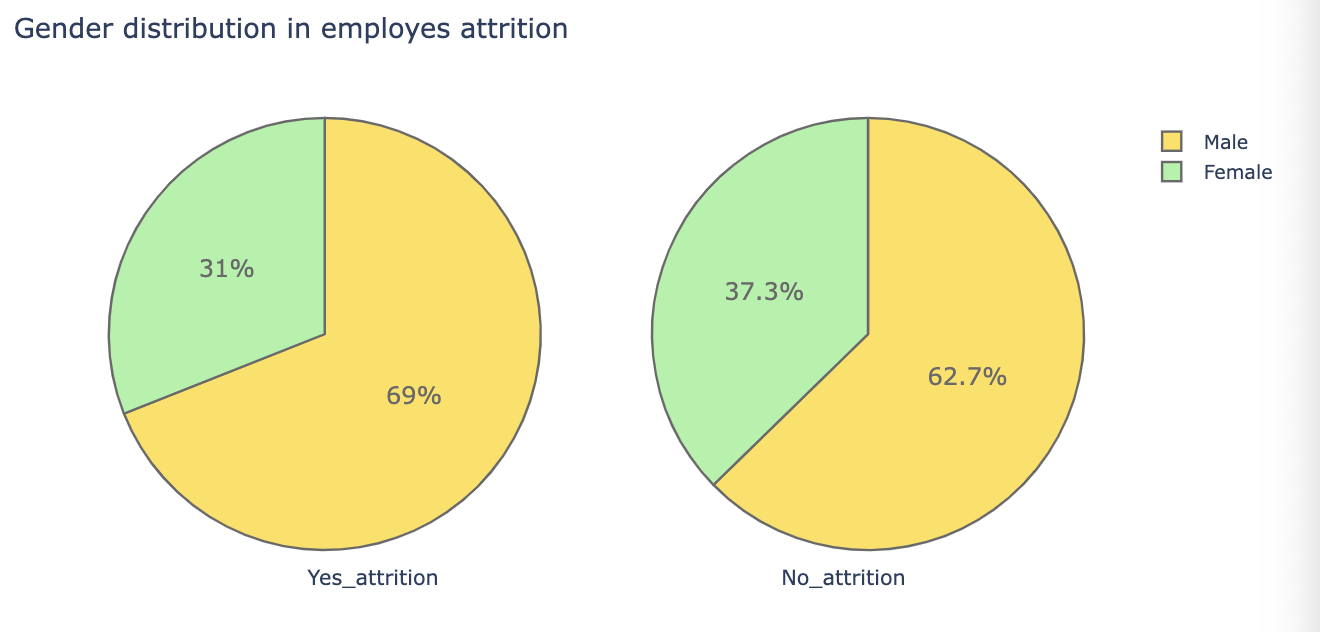
Refrence.
Plotly Tutorial - 파이썬 시각화의 끝판왕 마스터하기
Kaggle Series3 Episode 04
Binary Classification with a Tabular Credit Card Fraud Dataset
Modeling
RANDOM_STATE = 12
FOLDS = 5
STRATEGY = 'median'
TARGET = 'Class'
FEATURES = [col for col in train_data.columns if col != TARGET]LigtGBM Classifier
lgb_params = {
'objective' : 'binary',
'metric' : 'auc',
'device' : 'gpu',
}lgb_predictions = 0
lgb_scores = []
lgb_fimp = [] # feature importance
skf = StratifiedKFold(n_splits=FOLDS, shuffle=True, random_state=RANDOM_STATE)
for fold, (train_idx, val_idx) in enumerate(skf.split(train[FEATURES], train[TARGET])):
print('='*10, f'Fold={fold+1}', '='*10)
start_time = time.time()
X_train, X_valid = train.iloc[train_idx][FEATURES], train.iloc[val_idx][FEATURES]
y_train, y_valid = train.iloc[TARGET].iloc[train_idx], train[TARGET].iloc[val_idx]
model = LGBMClassifier(**lgb_params)
model.fit(X_train, y_train, verbose=0)
preds_valid = model.predict_proba(X_valid)[:, 1]
auc = roc_auc_score(y_valid, preds_valid)
lgb_scores.append(auc)
run_time = time.time() - start_time
print(f"Fold={fold+1}, AUC score: {auc:.2f}, Run Time: {run_time:.2f}s")
fim = pd.DataFrame(index=FEATURES,
data=model.feature_importances_,
columns=[f'{fold}_importance'])
lgb_fimp.append(fim)
test_preds = model.predict_proba(test_data[FEATURES])[:, 1]
lgb_predictions += test_preds/FOLDS
print("Mean AUC :", np.mean(lgb_scores))CatBoost
catb_params = {
"objective": "Logloss",
"iterations": 1000,
'l2_leaf_reg': 2.8,
"eval_metric": "AUC",
"random_seed": 12,
"task_type": "GPU"
}catb_predictions = 0
catb_scores = []
catb_fimp = []
skf = StratifiedKFold(n_splits=FOLDS, shuffle=True, random_state=RANDOM_STATE)
for fold, (train_idx, valid_idx) in enumerate(skf.split(train_data[FEATURES], train_data[TARGET])):
print(10*"=", f"Fold={fold+1}", 10*"=")
start_time = time.time()
X_train, X_valid = train_data.iloc[train_idx][FEATURES], train_data.iloc[valid_idx][FEATURES]
y_train , y_valid = train_data[TARGET].iloc[train_idx] , train_data[TARGET].iloc[valid_idx]
model = CatBoostClassifier(**catb_params)
model.fit(X_train, y_train, verbose=0)
preds_valid = model.predict_proba(X_valid)[:, 1]
auc = roc_auc_score(y_valid, preds_valid)
catb_scores.append(auc)
run_time = time.time() - start_time
print(f"Fold={fold+1}, AUC score: {auc:.2f}, Run Time: {run_time:.2f}s")
fim = pd.DataFrame(index=FEATURES,
data=model.feature_importances_,
columns=[f'{fold}_importance'])
catb_fimp.append(fim)
test_preds = model.predict_proba(test[FEATURES])[:, 1]
catb_predictions += test_preds/FOLDS
print("Mean AUC :", np.mean(catb_scores))XGBoost
xgb_params = {
"objective":"binary:logistic",
"eval_metric": "auc",
"n_estimators": 1000,
"random_state": 12,
'tree_method': 'gpu_hist',
'predictor': 'gpu_predictor',
}xgb_predictions = 0
xgb_scores = []
xgb_fimp = []
skf = StratifiedKFold(n_splits=FOLDS, shuffle=True, random_state=RANDOM_STATE)
for fold, (train_idx, valid_idx) in enumerate(skf.split(train_data[FEATURES], train_data[TARGET])):
print(10*"=", f"Fold={fold+1}", 10*"=")
start_time = time.time()
X_train, X_valid = train_data.iloc[train_idx][FEATURES], train_data.iloc[valid_idx][FEATURES]
y_train , y_valid = train_data[TARGET].iloc[train_idx] , train_data[TARGET].iloc[valid_idx]
model = XGBClassifier(**xgb_params)
model.fit(X_train, y_train,verbose=0)
preds_valid = model.predict_proba(X_valid)[:, 1]
auc = roc_auc_score(y_valid, preds_valid)
xgb_scores.append(auc)
run_time = time.time() - start_time
print(f"Fold={fold+1}, AUC score: {auc:.2f}, Run Time: {run_time:.2f}s")
test_preds = model.predict_proba(test_data[FEATURES])[:, 1]
fim = pd.DataFrame(index=FEATURES,
data=model.feature_importances_,
columns=[f'{fold}_importance'])
xgb_fimp.append(fim)
xgb_predictions += test_preds/FOLDS
print("Mean AUC :", np.mean(xgb_scores))Refrence.
Correlation tells the story
Use your Time wisely
[Playground S-3,E-4] 📊EDA + Modelling📈
Using the Original Dataset - Adversarial Validation
What I've learnt from the first four competitions this year
Kaggle Series3 Episode 07
Binary Classification with a Tabular Reservation Cancellation Dataset
EDA
plt.figure(figsize=(12,12))
plt.subplots_adjust(top=0.99, bottom=0.01, hspace=0.5, wspace=0.5)
x=1
for c in train_data.columns:
plt.subplot(6,3,x)
x += 1
sns.histplot(data=train_data, x=c, kde=True)
plt.show()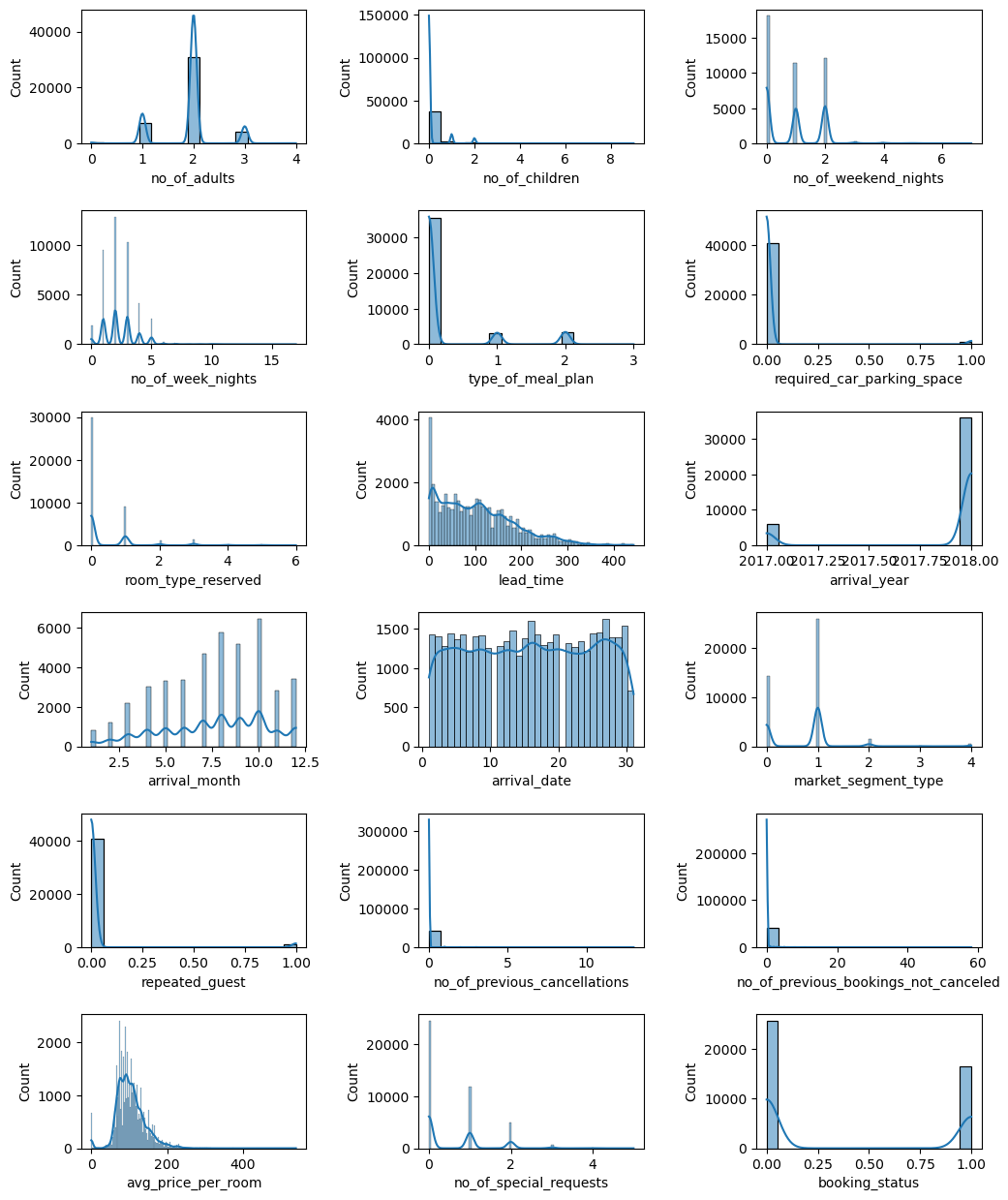
Feature Engineering
숙박 취소를 예측하는 데이터셋에서 Feature Engineering 진행한 것 중에 기억에 남는 부분.
arrival_year, arrival_month, arrival_date을 활용해서 분기, 봄여름가을겨울시즌, 공휴일 새 컬럼 생성해서 활용
df['arrival_week'] = df['arrival_full_date'].dt.isocalendar().week.astype(float)
df['arrival_dayofweek'] = df['arrival_full_date'].dt.dayofweek
df['arrival_quarter'] = df['arrival_full_date'].dt.quater
df['arrival_dayofyear'] = df['arrival_full_date'].dt.dayofyear
df['season'] = df.arrival_month*12 // 3 + 1
cal = USFederalHolidayCalendar()
holidays = cal.holidays(start='2017-01-01', end='2018-12-31')
df['is_holiday'] = 0
df.loc[df.arrival_full_date.isin(holidays), 'is_holiday'] = 1Refrence.
Fix your date anomalies like a boss
[PS S3E7, 2023] EDA and Submission
9th Place Solution XGB stack
Kaggle Series3 Episode 10
Binary Classification with a Tabular Pulsar Dataset
제대로 참여를 못해서 나중에 참고해서 시도해봐야지~ 하고 탭에만 추가해놨던 노트북들..
Refrence.
[PS S3E10, 2023] EDA and Submission
[9th Place] Model Predictions as Features
PS3E10 EDA| XGB/LGBM/CAT Ensemble Score 0.03153
Kaggle Series3 Episode 11
Regression with a Tabular Media Campaign Cost Dataset
제대로 참여를 못해서 나중에 참고해서 시도해봐야지~ 하고 탭에만 추가해놨던 노트북들..22
Collection of helpful ideas - S3E11
PSS3E11 Zoo of Models
A Framework for Tabular Regression - E16,14,11
(↘︎ EDA, 모델링 까지 하나의 프레임워크로 만들어놔서 쉽게 시작할수있어서 좋았었다..)
Kaggle Series3 Episode 12
Binary Classification with a Tabular Kidney Stone Prediction Dataset
EDA
from colorama import Style, Fore
rc = {
"axes.facecolor": "#FFF9ED",
"figure.facecolor": "#FFF9ED",
"axes.edgecolor": "#000000",
"grid.color": "#EBEBE7",
"font.family": "serif",
"axes.labelcolor": "#000000",
"xtick.color": "#000000",
"ytick.color": "#000000",
"grid.alpha": 0.4
}
sns.set(rc=rc)
from colorama import Style, Fore
red = Style.BRIGHT + Fore.RED
blu = Style.BRIGHT + Fore.BLUE
mgt = Style.BRIGHT + Fore.MAGENTA
gld = Style.BRIGHT + Fore.YELLOW
res = Style.RESET_ALLdef prepare_data(passes, origin):
train = pd.read_csv(passes + '/train.csv')
test = pd.read_csv(passes + '/test.csv')
original = pd.read_csv(origin)
sub = pd.read_csv(passes + '/sample_submission.csv')
if 'id' in train.columns:
train = train.drop(columns='id')
if 'id' in test.columns:
test = test.drop(columns='id')
print(f'{gld}[INFO] Shapes: '
f'{gld}\n[+] train -> {red}{train.shape}'
f'{gld}\n[+] test -> {red}{test.shape}'
f'{gld}\n[+] original -> {red}{original.shape}\n')
print(f'{gld}[INFO] Any missing values:'
f'{gld}\n[+] train -> {red}{train.isna().any().any()}'
f'{gld}\n[+] test -> {red}{test.isna().any().any()}'
f'{gld}\n[+] original -> {red}{original.isna().any().any()}')
return train, test, original, sub
흑백 세상에서 조금 더 눈에 띄게 결과를 볼 수 있어서 종종 애용..
Feature Distribution (boxplot)
train data, test data, original data 모두 사용할 때, 한번 두고 분포 보기 편했던 함수
def feature_distribution(origin=False):
features = test.columns
dm_train = train.copy()
dm_test = test.copy()
dm_original = original.copy()
dm_train['set'] = 'train'
dm_test['set'] = 'test'
if origin:
dm_original['set'] = 'original'
df_combined = pd.concat([dm_train, dm_test, dm_original])
else:
df_combined = pd.concat([dm_train, dm_test])
ncols = 2
nrows = np.ceil(len(features)/ncols).astype(int)
fig, axs = plt.subplots(ncols=ncols, nrows=nrows, figsize=(15, nrows*4))
for c, ax in zip(features, axs.flatten()):
sns.boxenplot(data=df_combined, x=c, ax=ax, y='set')
fig.suptitle('Distribution of features by set')
plt.tight_layout(rect=[0, 0, 1, 0.98])
plt.show()
Correlation
Feature 개수가 많을 때 반쪽만 시각화해서 조금 더 보기 편함
def plot_corr_heatmap(df: pd.core.frame.DataFrame, title_name: str='Train correlation') -> None:
corr = df.corr()
fig, axes = plt.subplots(figsize=(10, 5))
mask = np.zeros_like(corr)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(corr, mask=mask, linewidths=.5, cmap='YlOrBr_r', annot=True)
plt.title(title_name)
plt.show()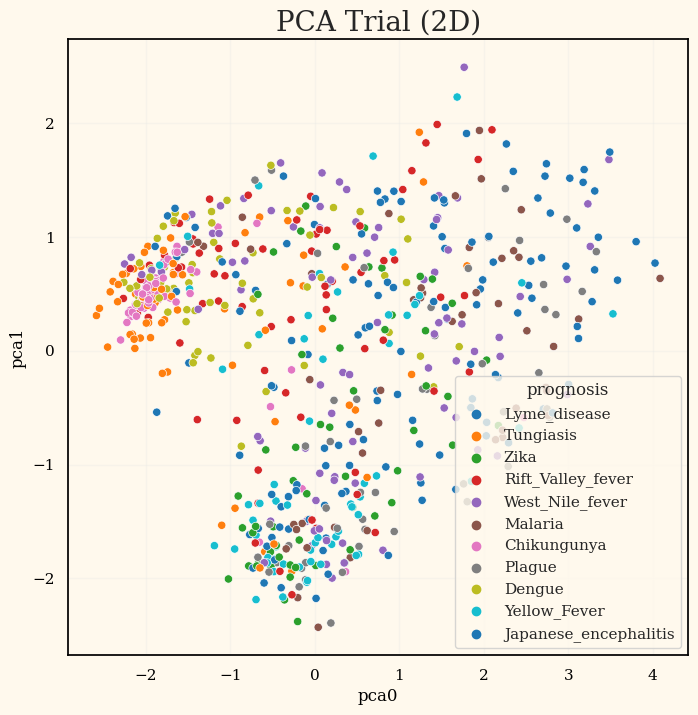
Kaggle Series3 Episode 13
Classification with a Tabular Vector Borne Disease Dataset
LabelEncoder
object 변수였던 Target 변수를 LabelEncoder()를 사용해서 변환
label_encoder = LabelEncoder()
encoded_labels = label_encoder.fit_transform(np.ravel(train['prognosis']]))
onehot_features = train.columns[:-1]
X_train, X_test, y_train, y_test = train_test_split(train[onehot_features],
encoded_labels, train_size=0.8, shuffle=True,
random_state=2, stratify=train[['prognosis']])
print(f'Size of X_train: {X_train.shape} Size of y_train: {y_train.shape}')
print(f'Size of X_test: {X_test.shape} Size of y_test: {y_test.shape}')PCA
64개의 feature들이 존재하고 그 중 데이터는 ~700개 정도로 차원축소 시도.
pca_trial = PCA(n_components=10)
pca_trial.set_output(transform='pandas')
X_transformed_pca = pca_trial.fit_transform(train[onehot_features])
display(X_transformed_pca.head())
evr = pca_trial.explained_variance_ratio_
for i in range(len(evr)):
print(f'PC_{i} variance explained ratio: {np.round(evr[i], decimals=3)}')
fig, ax = plt.subplots(figsize=(8,8))
sns.scatterplot(data=pd.concat([X_transformed_pca, train.prognosis], axis=1),
x='pca0', y='pca1', hue='prognosis', palette='tab10').set_title('PCA Trial (2D)', fontsize=20)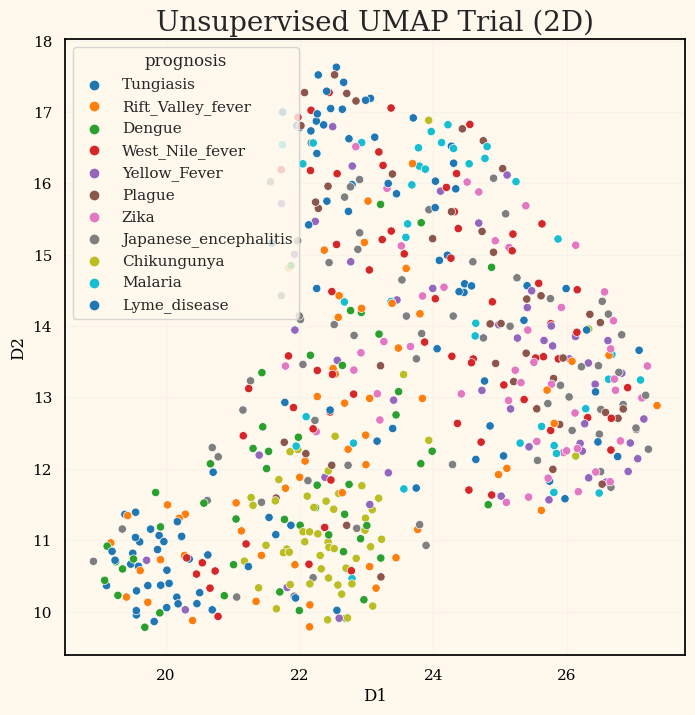
UMAP
umap_trial = umap.UMAP(n_neighbors=100, min_dist=0.5, n_components=2, target_weight=0)
X_transformed_umap = umap_trial.fit_transform(X=X_train)
plot_df = pd.concat([pd.DataFrame(X_transformed_umap, columns=['D1','D2']),
pd.DataFrame({'prognosis': label_encoder.inverse_transform(y_train)})], axis=1)
display(plot_df.head())
fig, ax = plt.subplots(figsize=(8,8))
ax = np.ravel(ax)
sns.scatterplot(data=plot_df, x='D1', y='D2', hue='prognosis', palette='tab10', ax=ax[0]).set_title('Unsupervised UMAP Trial (2D)', fontsize=20)대회 자체 평가지표인 MAP@3를 기준으로 비교했을 때,
PCA 후 테스트한 결과가 이전 결과보다 0.03정도 좋아지긴했었다.
UMAP은 오히려 안좋았었는데, 모두 사용해서 평가 내본 결과는 없어서 아쉽다.
Refrence.
😷simple EDA and multi-class classification
PS3E13 EDA| Decomposition+Ensemble&RankPredict
S3.E13 | EDA+PCA+XGB w/ Optuna
전체적으로 하다만것같은 에피소드들이 너무 많아서 다음 에피소드나 과제부터는 가설이나 어떤 부분을 찾아가고싶은지 생각해보면서 분석을 해야할것같다.
데이터있다고 냅다 분석해야지! 하고 아무생각없이 들어간게 많았는데,
오히려 갈피를 못잡고 그냥 하던거 따라적는 부분만 늘어갔던것같아서 새로운 인사이트에 대해 고민하면서 해나가야할 것 같다.