setwd("c:/Rwork")
subjects <- read.csv("dataset2/subjects.csv", head=T)
head(subjects, 3)
tail(subjects, 3)
library(psych)
options(digits=3)
(corMat <- cor(subjects))
library("GPArotation") # 사교회전(“oblimin”옵션)의 수행에 필요함
EFA <- fa(r = corMat, nfactors = 2, rotate="oblimin", fm = "pa")
EFA
ls(EFA)
EFA$loadings
load<-EFA$loadings[,1:2]
plot(load, type="n")
text(load, labels=names(subjects), cex=.7)
library(nFactors)
ev <- eigen(cor(subjects)) # get eigenvalues
ap <- parallel(subject=nrow(subjects),var=ncol(subjects), rep=100,cent=.05)
nS <- nScree(x=ev$values, aparallel=ap$eigen$qevpea)
plotnScree(nS)

library(FactoMineR)
result <- PCA(subjects)
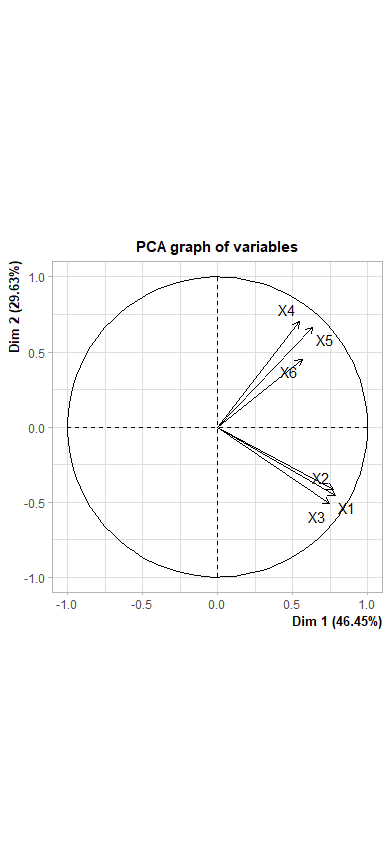
names(subjects) <-c("X1", "X2", "X3", "X4", "X5", "X6")
names(subjects)
mydata.cov <- cov(subjects)
model.mydata <- specifyModel()
mydata.sem <- sem(model.mydata, mydata.cov, nrow(subjects))
summary(mydata.sem)
stdCoef(mydata.sem)


