data("iris")
head(iris)
cor(iris[1:4])
iris2 <- iris[, 1:4]
ir.species <- iris[,5]
prcomp.result2 <- prcomp(iris2, center=T, scale=T)
prcomp.result2
summary(prcomp.result2)
plot(prcomp.result2, type="l")
prcomp.result2$rotation
iris2
Result3 <- as.matrix(iris2) %*% prcomp.result2$rotation
head(Result3)
final2 <- cbind(ir.species, as.data.frame(Result3))
final2
final2[,1] <- as.factor(final2[,1])
colnames(final2)[1] <- "label1"
final2
fit3 <- lm(label1 ~ PC1 + PC2, data=final2)
fit3_pred <-predict(fit3, newdata=final2)
b2 <- round(fit3_pred)
a2 <- ir.species
table(b2,a2)
data(iris)
head(iris, 3)
log.ir <- log(iris[, 1:4])
ir.species <- iris[, 5]
ir.pca <- prcomp(log.ir, center = TRUE, scale. = TRUE)
print(ir.pca)
plot(ir.pca, type = "l")
summary(ir.pca)
predict(ir.pca, newdata=tail(log.ir, 2))
biplot(ir.pca)
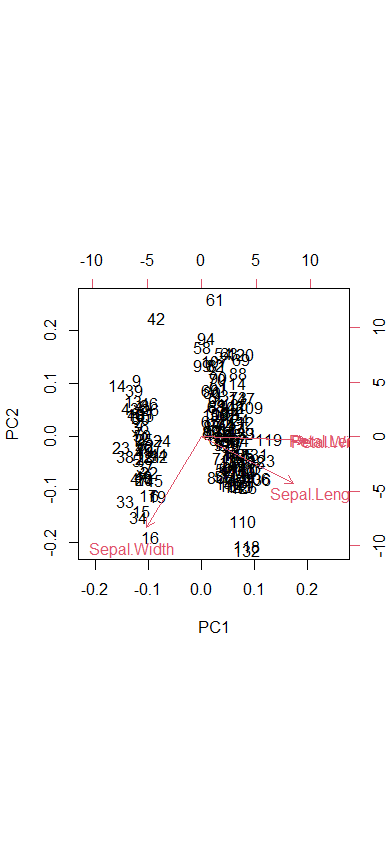
library(devtools)
library(ggbiplot)
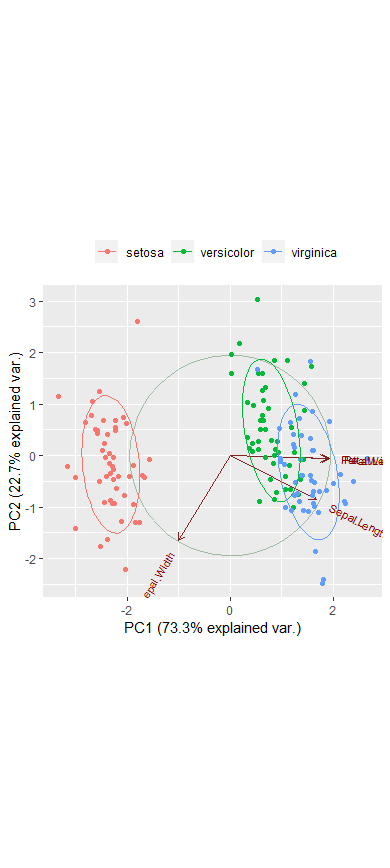
g <- ggbiplot(ir.pca, obs.scale=1, var.scale=1, groups=ir.species, ellipse=TRUE, circle=TRUE)
g <- g + scale_color_discrete(name='')
g <- g + theme(legend.direction='horizontal', legend.position='top')
print(g)
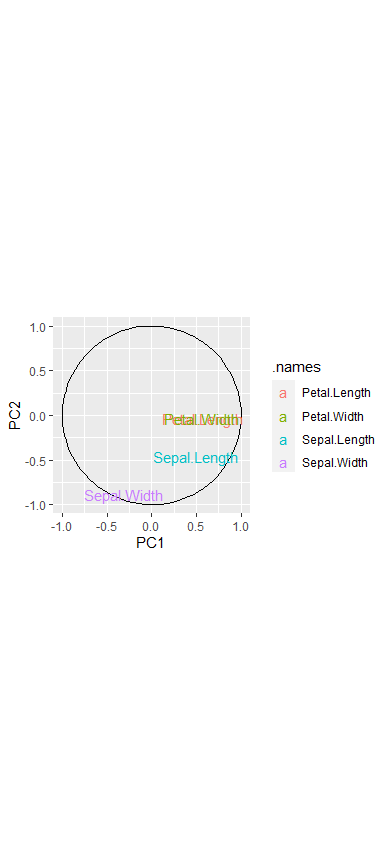
require(ggplot2)
theta <- seq(0,2*pi,length.out = 100)
circle <- data.frame(x = cos(theta), y = sin(theta))
p <- ggplot(circle,aes(x,y)) + geom_path()
loadings <- data.frame(ir.pca$rotation, .names = row.names(ir.pca$rotation))
p + geom_text(data=loadings,
mapping=aes(x = PC1, y = PC2, label = .names, colour = .names)) +
coord_fixed(ratio=1) +labs(x = "PC1", y = "PC2")
require(caret)
trans = preProcess(iris[,1:4], method=c("BoxCox", "center", "scale", "pca"))
PC = predict(trans, iris[,1:4])
head(PC, 3)
trans$rotation



