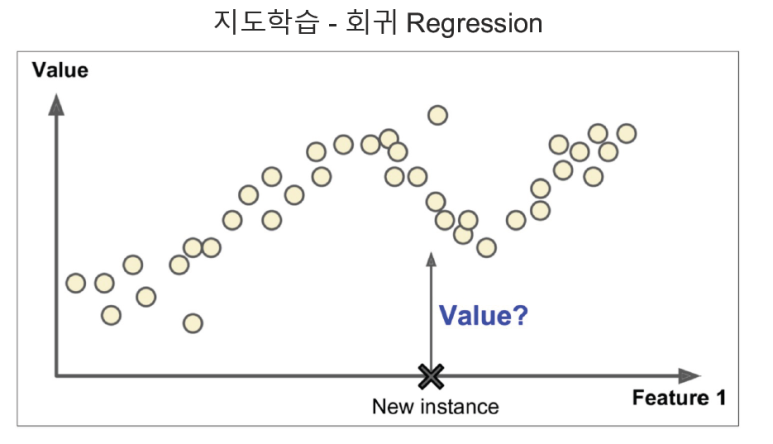
해당 글은 제로베이스데이터스쿨 학습자료를 참고하여 작성되었습니다
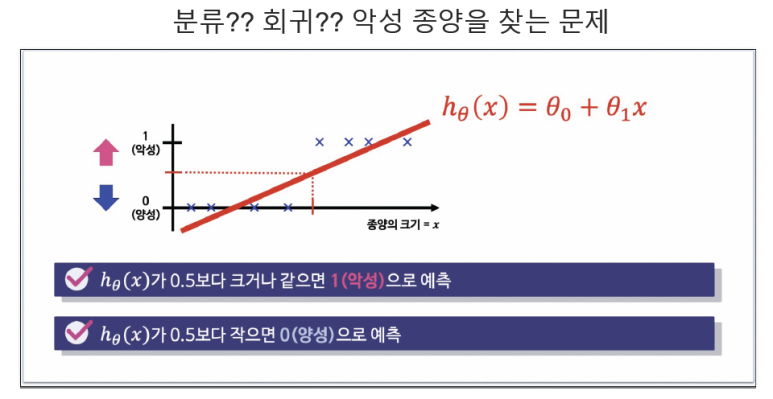
1. Logistic Regression
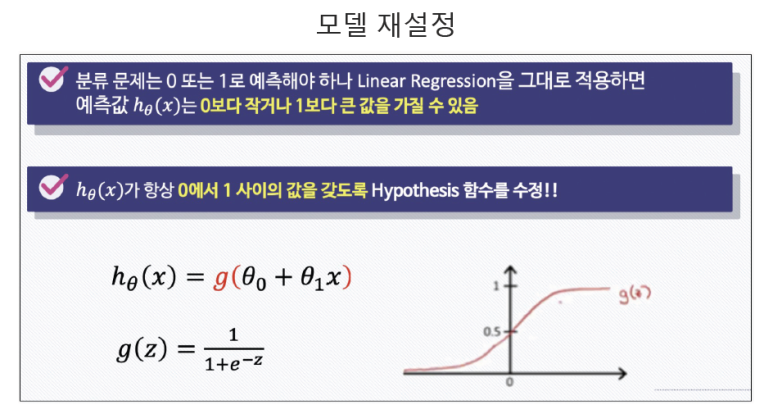
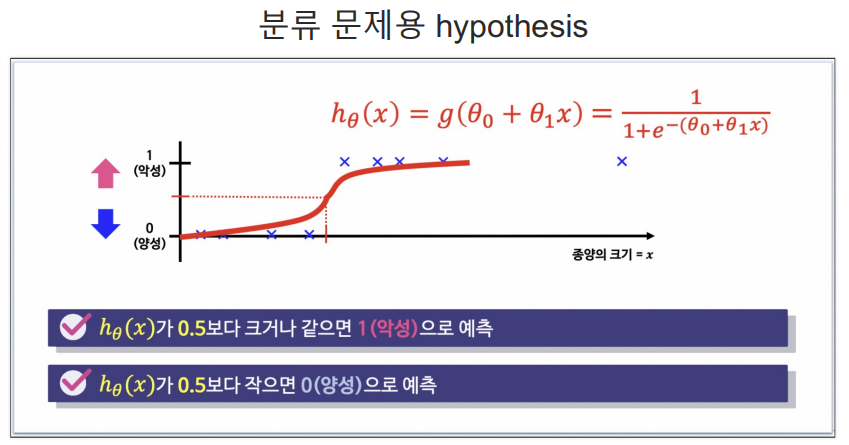
- 데이터의 스케일을 시그모이드 함수를 이용하여 0~1 사이로 스케일 설정
- 특정기준으로 데이터를 이진분류(0 or 1)로 결정하는 회귀방식
- ex) 확률이 0.5보다 크면 악성(1), 낮으면 양성(0)

시그모이드 실습
시그모이드 함수로 데이터를 0~1 사이로 스케일 설정
import numpy as np
import matplotlib.pyplot as plt
z = np.arange(-10, 10, 0.01)
g = 1 / (1+np.exp(-z))
plt.plot(z,g)
plt.show()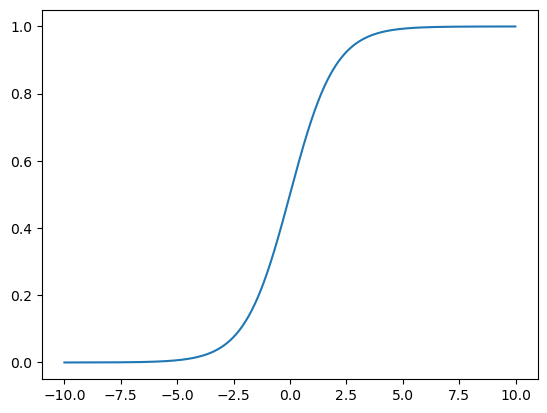
축 옮기기
plt.figure(figsize=(12,8))
ax = plt.gca()
ax.plot(z,g)
ax.spines['left'].set_position('zero')
ax.spines['bottom'].set_position('center')
ax.spines['right'].set_color('none')
ax.spines['top'].set_color('none')
plt.grid()
plt.show()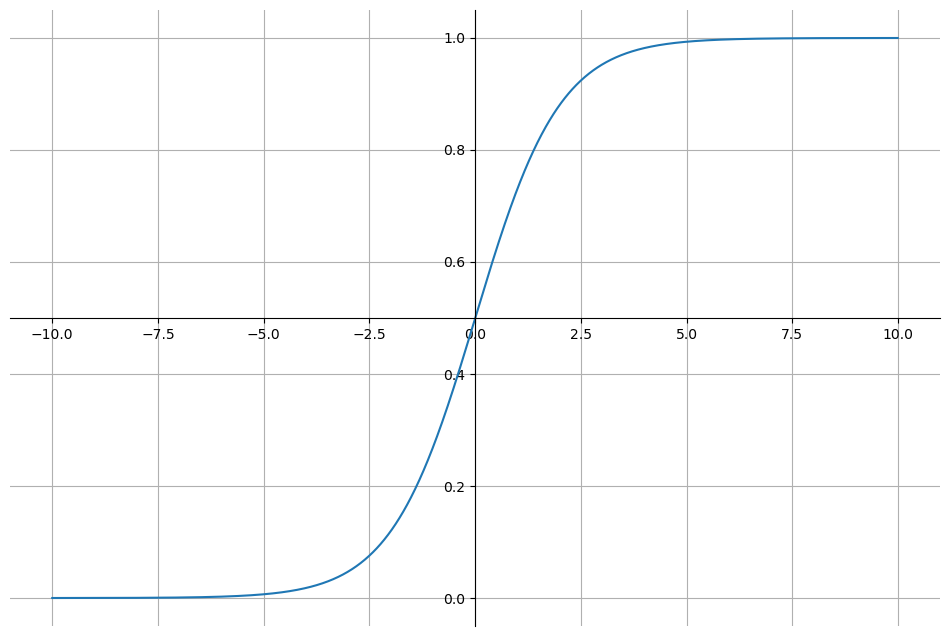
와인데이터 실습
와인데이터 가져오기
import pandas as pd
wine_url = 'https://raw.githubusercontent.com/Pinkwink/ML_tutorial/master/dataset/wine.csv'
wine = pd.read_csv(wine_url, index_col=0)
wine['taste'] = [1. if grade>5 else 0. for grade in wine['quality']]
X = wine.drop(['taste', 'quality'], axis=1)
y = wine['taste']Logistic Regression 모델 학습
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=13)
estimators = [('scaler', StandardScaler()),
('clf', LogisticRegression(solver='liblinear', random_state=13))]
pipe = Pipeline(estimators)
pipe.fit(X_train, y_train)
y_pred_tr = pipe.predict(X_train)
y_pred_test = pipe.predict(X_test)
print('Train Acc : ', accuracy_score(y_train, y_pred_tr))
print('Test Acc : ', accuracy_score(y_test, y_pred_test))
-----------------------------------
Train Acc : 0.7444679622859341
Test Acc : 0.7469230769230769Decision Tree 모델 학습
from sklearn.tree import DecisionTreeClassifier
wine_tree = DecisionTreeClassifier(max_depth=2, random_state=13)
wine_tree.fit(X_train, y_train)
models = {'Logistic Regression' : pipe,
'Decision Tree' : wine_tree}AUC 그래프를 이용한 모델간 비교
- 로지스틱 회귀가 의사결정나무보다 성능이 좋음
from sklearn.metrics import roc_curve
import matplotlib.pyplot as plt
plt.figure(figsize=(10,8))
plt.plot([0,1], [0,1])
for model_name, model in models.items():
pred = model.predict_proba(X_test)[:, 1]
fpr, tpr, thresholds = roc_curve(y_test, pred)
plt.plot(fpr, tpr, label = model_name)
plt.grid()
plt.legend()
plt.show()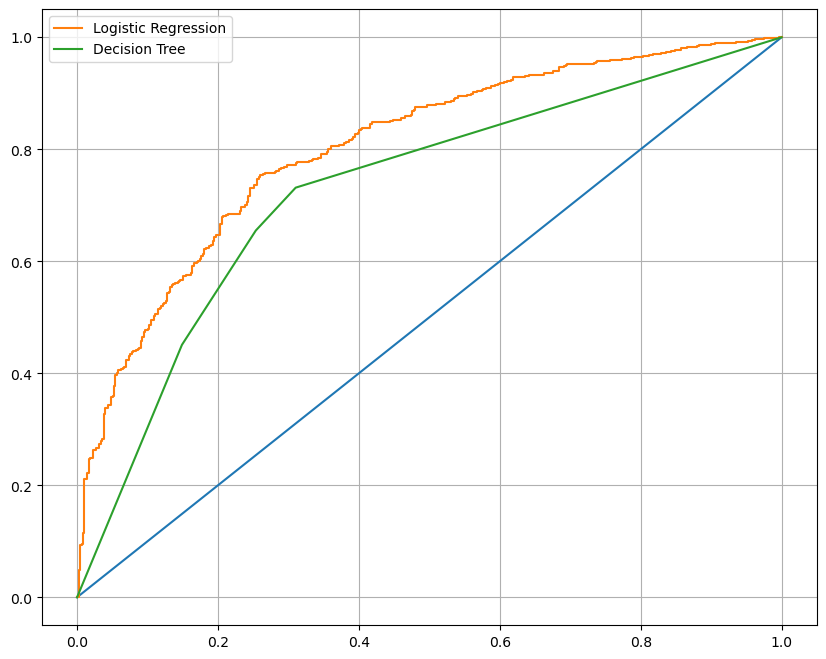
2. PIMA 인디언 당뇨병 예측
개요
- 50년대까지 PIMA인디언은 당뇨가 없었음
- 20세기 말, 50%가 당뇨에 걸림
- 50년만에 50%의 인구가 당뇨에 걸림
목표
- PIMA인디언의 당뇨병 예측하기
절차
- 1) 데이터 이해
- 2) 데이터 전처리
- 3) 모델 학습
- 4) 데이터 시각화
- 5) 해석
1) 데이터 이해
# 인디언 데이터 가져오기
import pandas as pd
PIMA_url = 'https://raw.githubusercontent.com/Pinkwink/ML_tutorial/master/dataset/diabetes.csv'
PIMA = pd.read_csv(PIMA_url)
PIMA.head()

2) 데이터 전처리
데이터타입 float로 통일
PIMA = PIMA.astype('float')
PIMA.info()
-------------------------------------------------------
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null float64
1 Glucose 768 non-null float64
2 BloodPressure 768 non-null float64
3 SkinThickness 768 non-null float64
4 Insulin 768 non-null float64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null float64
8 Outcome 768 non-null float64
dtypes: float64(9)
memory usage: 54.1 KB상관관계확인
- Outcome과 상관계수가 0.2가 넘는 것이 4개가 있음
import seaborn as sns
import matplotlib.pyplot as plt
plt.figure(figsize=(12,10))
sns.heatmap(PIMA.corr(), annot=True, cmap='YlGnBu')
plt.show()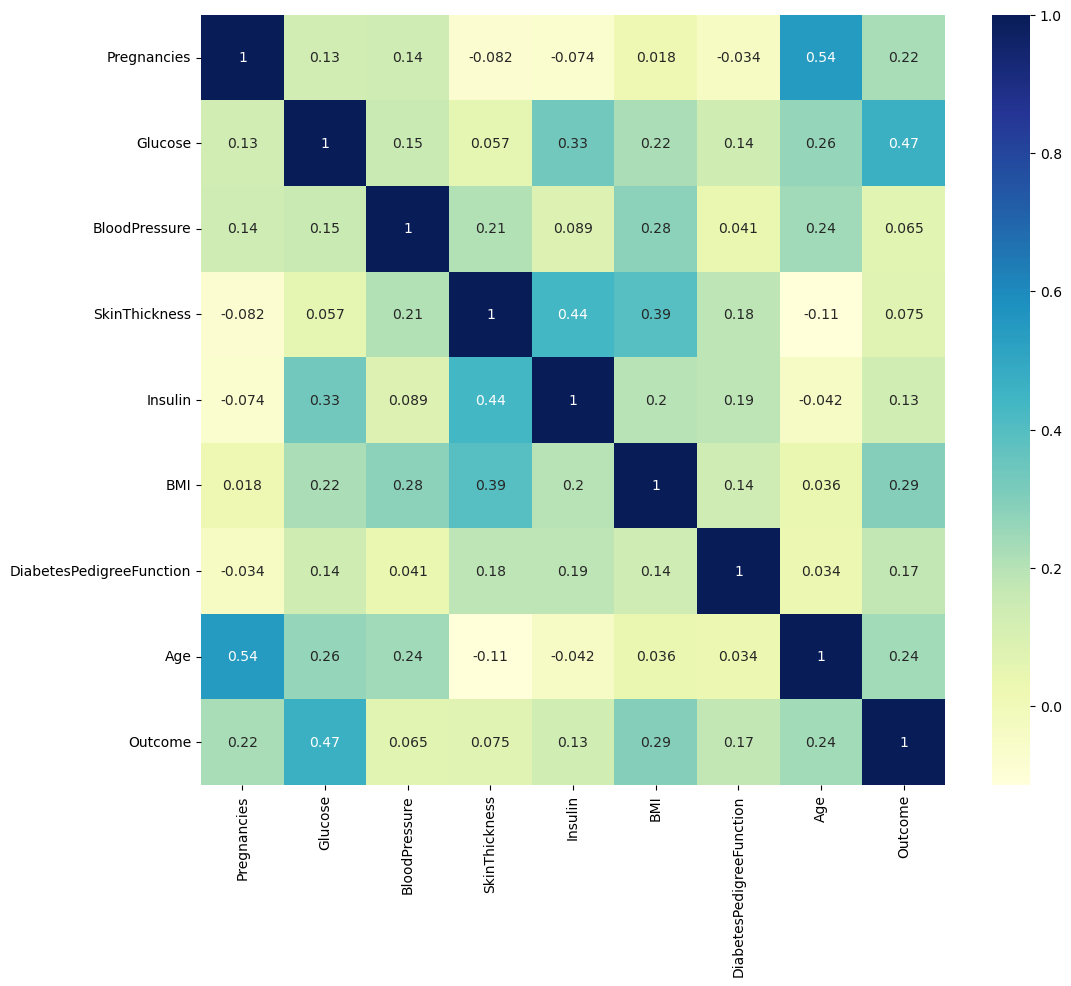
논리결측값
- 결측치는 아니지만 값이 0인 데이터
(PIMA==0).astype(int).sum()
---------------------------------
Pregnancies 111
Glucose 5
BloodPressure 35
SkinThickness 227
Insulin 374
BMI 11
DiabetesPedigreeFunction 0
Age 0
Outcome 500
dtype: int64- 평균으로 대체
- 인슐린 데이터는 배제
zero_features = ['Glucose','BloodPressure','SkinThickness','BMI']
PIMA[zero_features] = PIMA[zero_features].replace(0, PIMA[zero_features].mean())
(PIMA==0).astype(int).sum()
---------------------------------------
Pregnancies 111
Glucose 0
BloodPressure 0
SkinThickness 0
Insulin 374
BMI 0
DiabetesPedigreeFunction 0
Age 0
Outcome 500
dtype: int643) 모델 학습
from sklearn.model_selection import train_test_split
X = PIMA.drop(['Outcome'], axis=1)
y = PIMA['Outcome']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=13, stratify=y)
estimators = [('scaler', StandardScaler()),
('clf', LogisticRegression(solver='liblinear', random_state=13))]
pipe_lr = Pipeline(estimators)
pipe_lr.fit(X_train, y_train)
pred = pipe_lr.predict(X_test)평가
from sklearn.metrics import accuracy_score, recall_score, precision_score
from sklearn.metrics import roc_auc_score, f1_score
print('Accuracy : ', accuracy_score(y_test, pred))
print('Recall : ', recall_score(y_test, pred))
print('Precision : ', precision_score(y_test, pred))
print('AUC score : ', roc_auc_score(y_test, pred))
print('F1 score : ', f1_score(y_test, pred))
-------------------------------------
Accuracy : 0.7727272727272727
Recall : 0.6111111111111112
Precision : 0.7021276595744681
AUC score : 0.7355555555555556
F1 score : 0.6534653465346535classification_report
- precision : 양성으로 예측한 것 중에 양성인 비율
- recall : 양성 데이터 중에 양성으로 예측한 비율
- f1-score : precision과 recall을 합친 지표
- support : 데이터 갯수
- marcro avg : 각 지표의 평균
- weigthed avg : 클래스 분포가 적용된 각 지표의 평균
- ex) weigthed avg(precision)
- ex) weigthed avg(precision)
from sklearn.metrics import classification_report
print(classification_report(y_test, pred))
------------------------------------------------------
precision recall f1-score support
0.0 0.80 0.86 0.83 100
1.0 0.70 0.61 0.65 54
accuracy 0.77 154
macro avg 0.75 0.74 0.74 154
weighted avg 0.77 0.77 0.77 154confusion_matrix
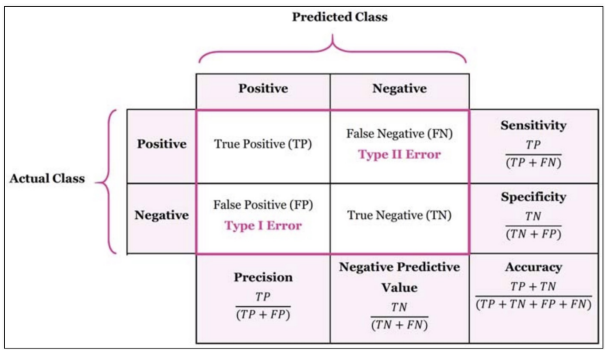
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, pred)
--------------------------
array([[86, 14],
[21, 33]], dtype=int64)precision_recall_curve
- threshold에 따른 precision과 recall의 변화
from sklearn.metrics import precision_recall_curve
plt.figure(figsize=(10,8))
precisions, recalls, thresholds = precision_recall_curve(y_test, pred)
plt.plot(thresholds, precisions[:len(thresholds)], label='precision')
plt.plot(thresholds, recalls[:len(thresholds)], label='recall')
plt.grid()
plt.legend()
plt.show()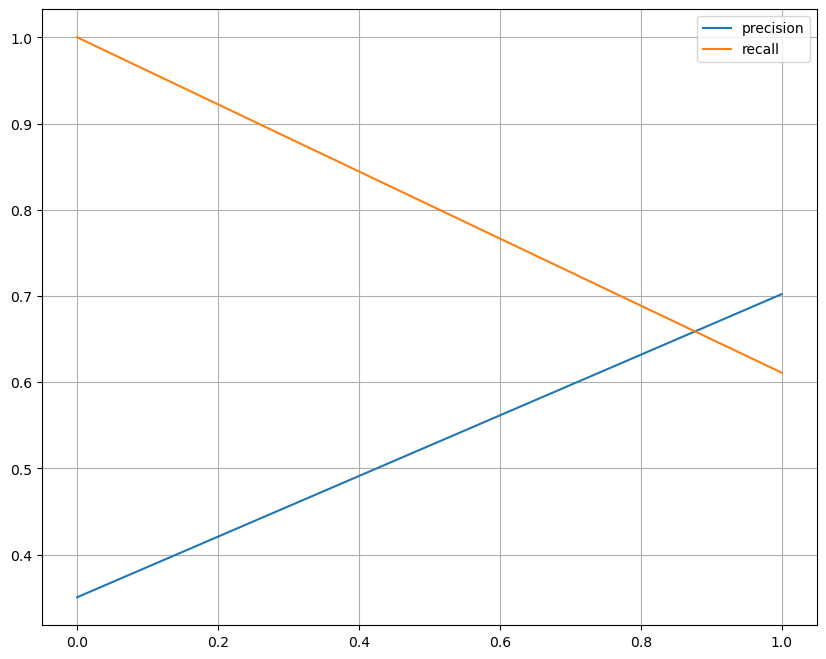
각 계수 값 확인
import pandas as pd
coeff = list(pipe_lr['clf'].coef_[0])
labels = list(X_train.columns)
features = pd.DataFrame({'Features':labels, 'importance':coeff}).sort_values(by='importance', ascending=False)
features
----------------------------------------
Features importance
1 Glucose 1.201424
5 BMI 0.620405
6 DiabetesPedigreeFunction 0.366694
0 Pregnancies 0.354266
7 Age 0.171960
3 SkinThickness 0.033947
2 BloodPressure -0.158401
4 Insulin -0.1628654) 데이터 시각화
features = pd.DataFrame({'Features':labels, 'importance':coeff})
features.sort_values(by=['importance'], ascending=True, inplace=True)
features['positive'] = features['importance'] > 0
features.set_index('Features', inplace=True)
features['importance'].plot(kind='barh',figsize=(11,6),
color=features['positive'].map({True:'blue', False:'red'}))
plt.xlabel('Importance')
plt.show()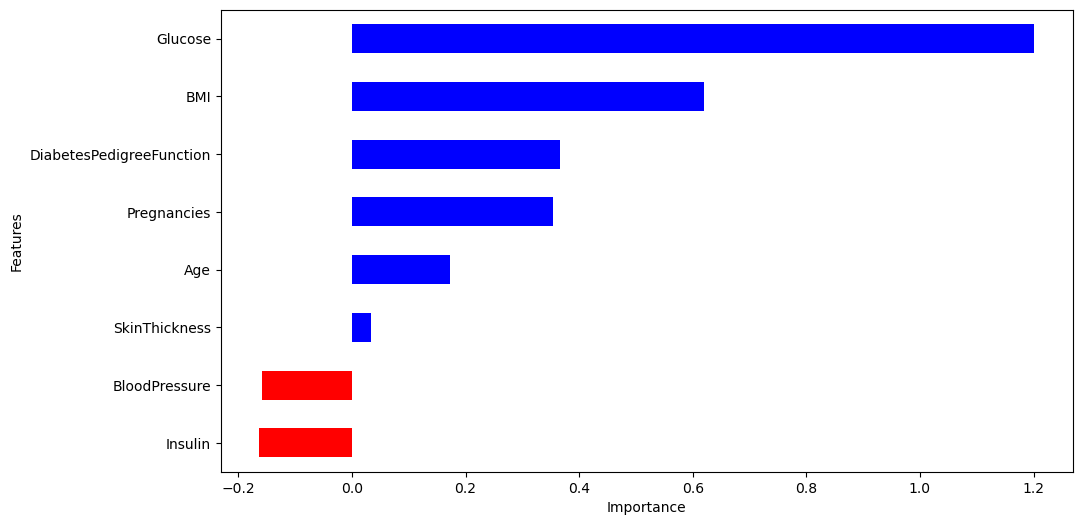
5) 해석
-
Glucose(포도당 부하 검사 수치)와 BMI가 당뇨에 큰 영향을 미친다.
-
혈압은 예측에 부정적 영향을 준다.
