What is Graph Algorithms?
Graph G = ( V, E )
- set V of n vertices.
- set E of m edges between vertices.
Degree d(V) of vertex v
- The number of vertices connected by edge to V.
- Σd(v) = 2m
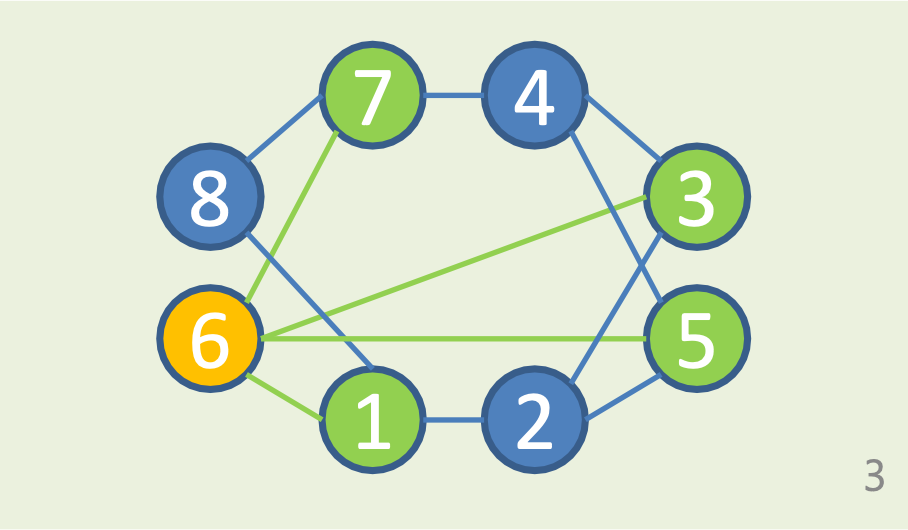
Edge 기준: 1 → 2와 2 → 1은 서로 같음.
d(V) 기준: 1 → 2와 2 → 1은 서로 기준(출발점)이 다르기 때문에 개별로 취급!즉,Edge는 시작점과 끝나는 점의 순서 상관없이 하나로 취급하기 때문에 총합 d(V)의 1/2!
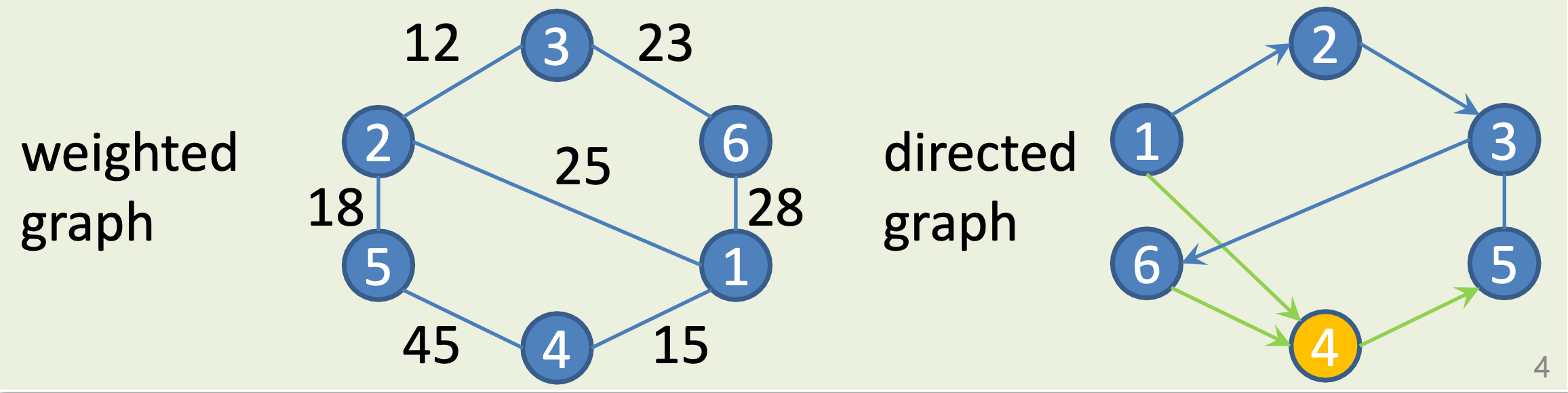
Weighted and Directed Graphs
-
Weighted Graph: edges have weights.
-
Directed graph: arcs point from vertex to other.
- indegree of V = the number of incoming arcs.
- outdegree of V = the number of outgoing arcs.
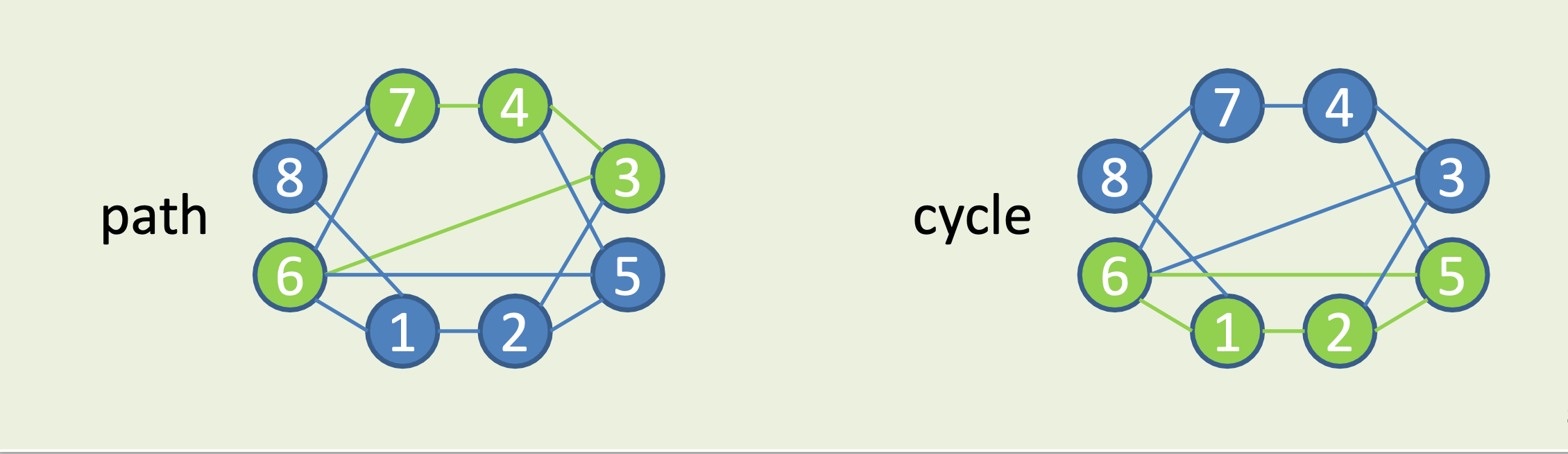
Paths and Cycles
-
Path: continous sequence of edges, no repeated vertices allowed.
-
Cycle: (=closed path) path starting and ending at the same vertex.
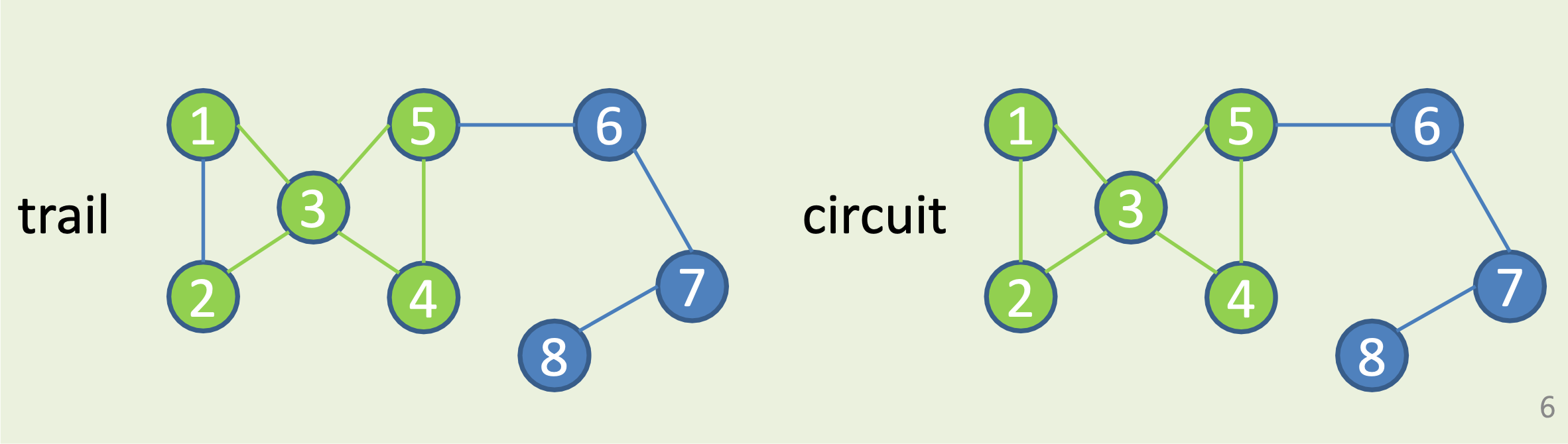
Trails and Circuits
-
Trail: continuous sequence of edges, no repeated edges allowed.
-
Circuit: closed trail = trail starting and ending in same vertex.
Connected graphs & complete graphs
-
Connected: Graph 𝐺 exists a path between every pair of vertices.
-
Disconnected: Graph 𝐺 is not connected; 𝐺 can be partitioned into connected components.
-
Complete: Graph 𝐺 is complete if there is an edge between every two vertices.
Example: Knight's puzzles and Hamiltonian graphs
1. Knight's Puzzle
Given: WWBB configuration
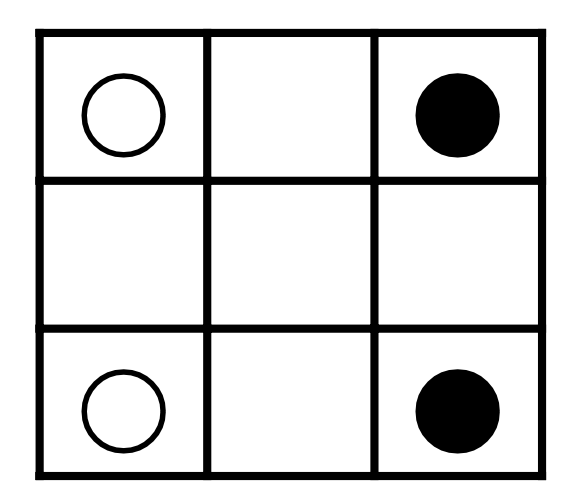
- 3 x 3 chessboard
- 2 white and 2 black knights in corners
- Knights ONLY allows to move
- 2 rows + 3 columns
- 3 rows + 2 columns
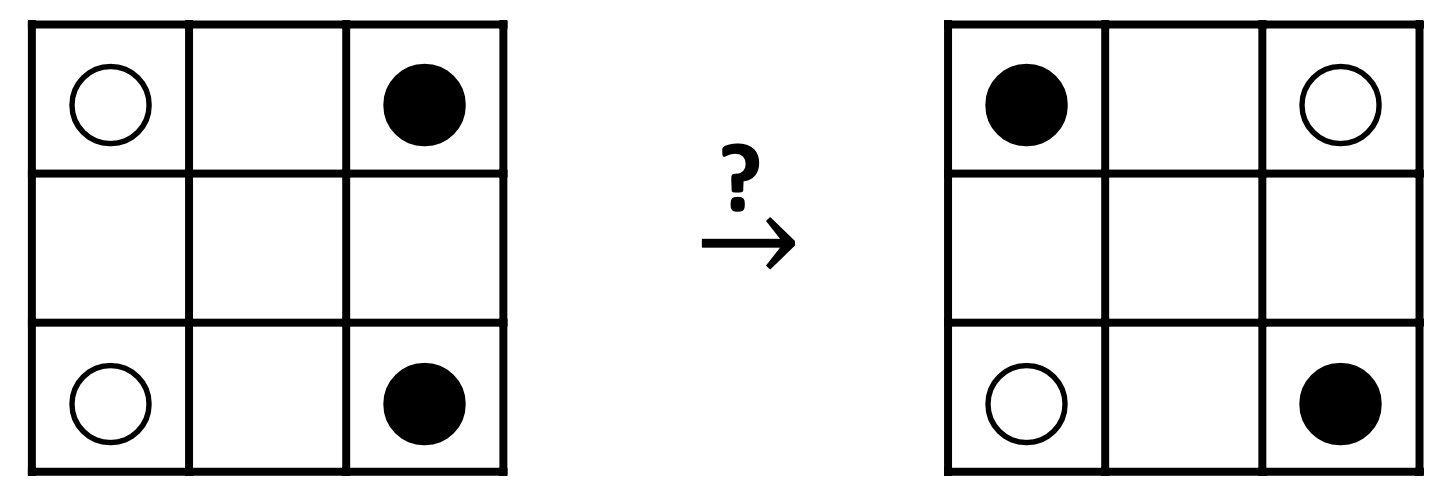
Question: can knights move from WWBB to BWBW
configuration?
s.t. never two knights on one square
-
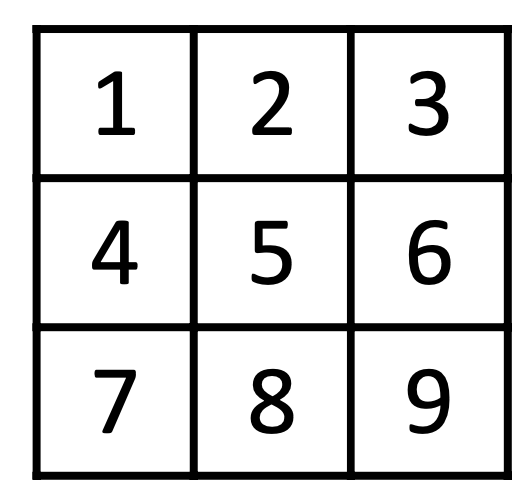
Number the Squares
-
Corresponding graph
- vertices are squares.
- edge between vertices if possible to move between squares.
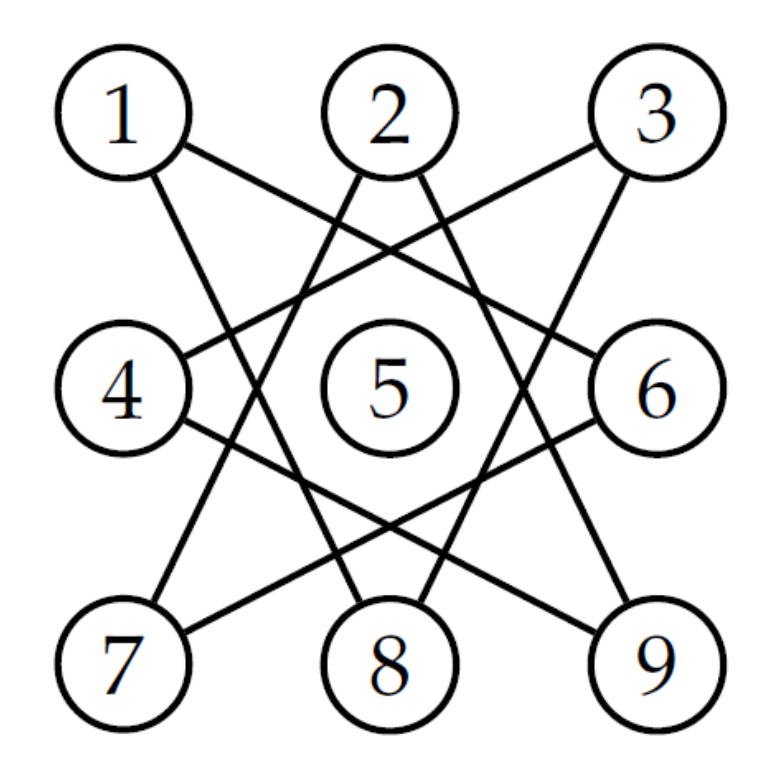
- Graph Rearranged
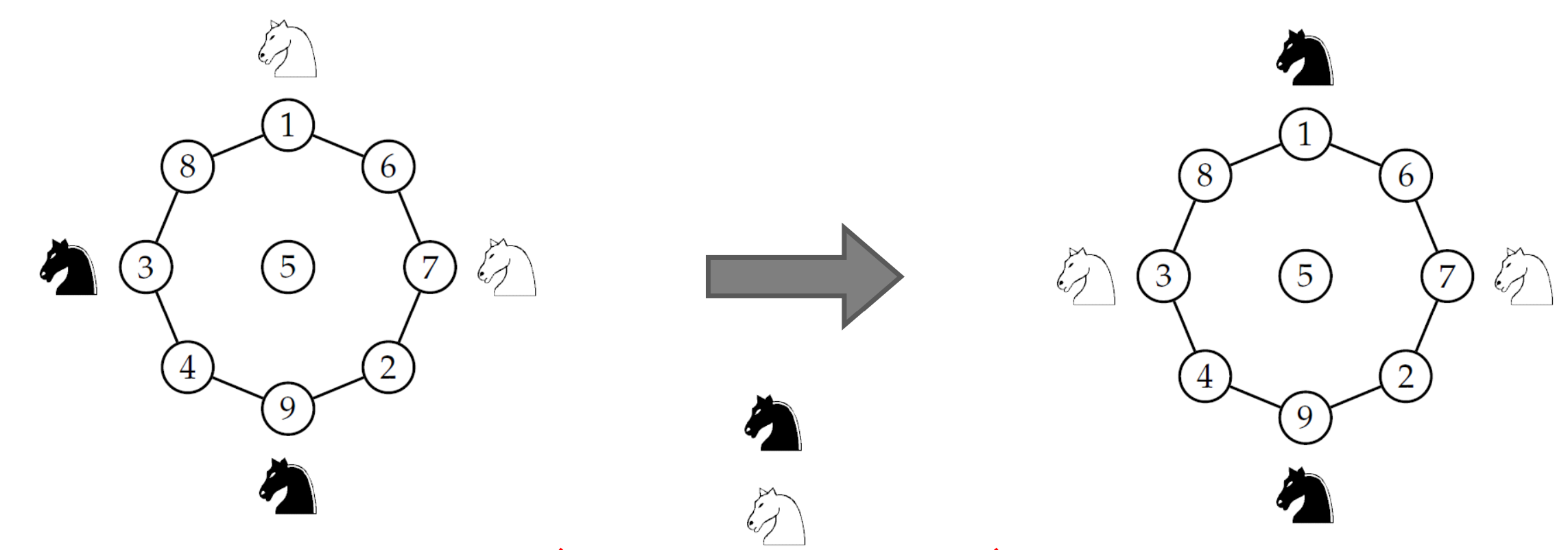
-
Knights들은 위의 graph를 기준으로 clockwise or anti-clockwise 방향으로만 움직일 수 있음.
-
따라서, knights cannot move from WWBB to BWBW
configuration without violating constraint
(우리가 원하는 결과를 얻으려면, 하나의 자리에 두개의 knight가 있거나, jump해서 자리를 이동해야만 한다.)
2. Hamilton's puzzle: Around the world
-
Hamilton's Graph: a closed loop on a graph where every node(VERTEX) is visited exactly only once.
-
Is there Hamiltonian algorithm?
- NO efficient algorithm is known/exist.
- Intractable problem (다루기 힘든 문제)
Related Problem
→ Traveling Salesman Problem(TSP)
- in (complete) weighted graph
- vertices: cities
- edges: distances
- find Hamiltonian Cycle of minimum weight
3. Eulerian graphs
Definition of Eulerian Circuit
A circuit which transverses every edge in a graph exactly once.
(즉, 같은 vertex로 돌아와도 상관없음.)
- Hamilton은 every vertex임
Property
Graph is Eulerian if and only if every vertex has an even degree.
(하나의 꼭짓점 당 두개의 꼭짓점만 이어져야 한다.)
STEPS
-
select an arbitrary vertex V.
-
construct an arbitrary circuit by traversing unused edges, til we end up in V again.
-
if the constructed circuit is not Eulerian, then circuit must contain a vertex W that still has untraversed edges.
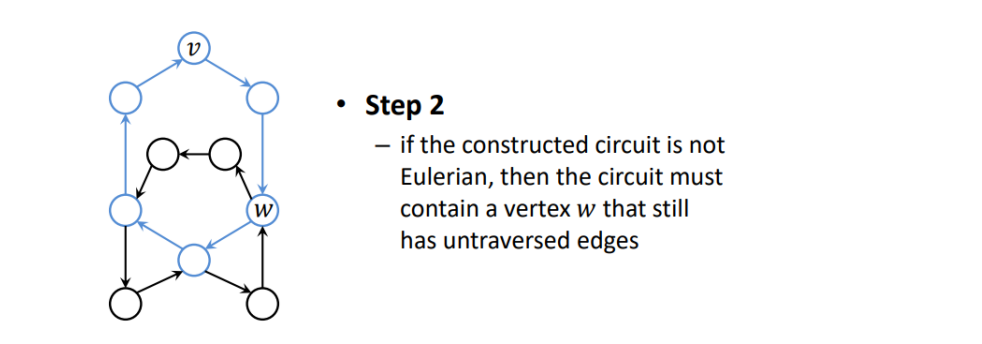
-
use W to construct another circuit using the untraversed edges.
-
combine the two circuits
- go from V to W
- go from W back to W
- go from W back to V

DNA Sequencing
Shortest superstring problem (SSP)
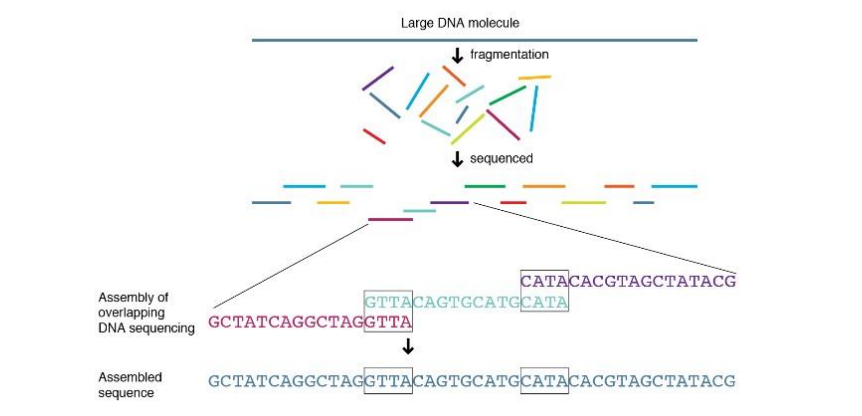
-
DNA sequence를 여러조각으로 쪼갠 후(fragmentation) 각 조각들의 염기서열을 분석한 후 다시 assembly하는 과정.
-
Overlapping된 염기서열도 존재함.
How to solve
-
Define "Overlap" function overlap(Si, Sj)
→ length of longest perfix of Sj that matches with a suffix of Si -
Transform SSP Problem to TSP problem
→ complete directed graph with n verices
→ vertex i is substring Si
→ weight on edge(i, j) is → overlap(Si, Sj) -
TSP = Hamiltonian path problem
-
Greedy Approach
→ Repeatedly merge pair of substrings with maximum overlap until one string remains.
DNA Arrays: sequencing by hybridization(using Hamiltonian path)
- For unknown DNA sequence
- no information about position in DNA sequence
- SSP and SBH very similar as algorithmic problems
- SBH is special case of SSP
→ strings Si have fixed length
(SSP: Shortcut Superstring Problem / SBH: Sequencing By Hybridization)
How to solve
-
Overlap of two l-mers p and q
→ p and q overlap(p,q) = l-1
(= last l-1 chars of p coincide with first l-1 chars of q) -
Given Spectrum(s,l) for unknown s, construct directed graph
→ vertices are l-mers in spectrum(s,l)
→ arc from p to q if p and q overlap
-
Then one-to-one correspondence between
→ paths that visit every vertex in graph exactly once
→ DNA seqeunces with given spectrum
→ Hamiltonian Path : paths that visit every vertex in graph exactly once.
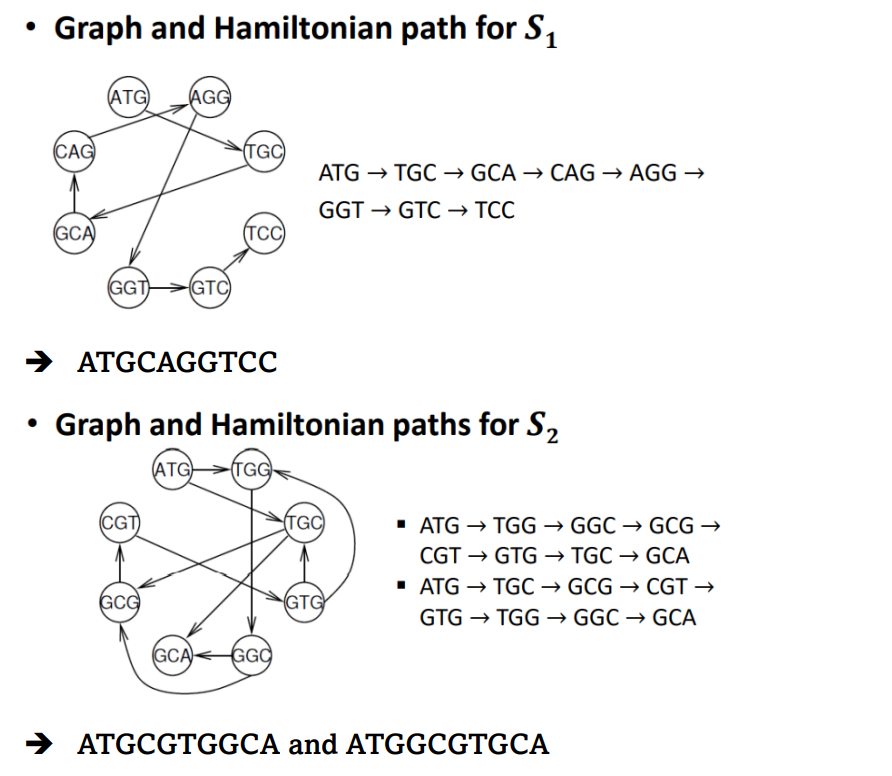
< Example >
1. S1 = { ATG, AGG, TGC, TCC, GTC, GGT, GCA, CAG}
2. S2 = { ATG, TGG, TGC, GTG, GGC, GCA, GCG. CGT}
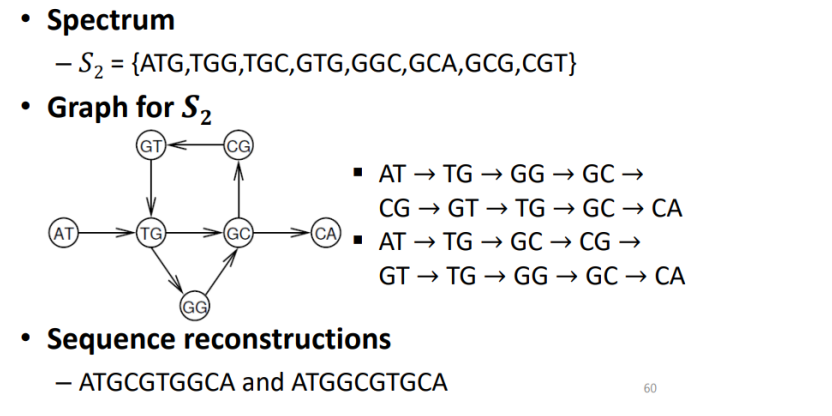
DNA Arrays: sequencing by hybridization(using Eulerian trail)
- construct Graph where EDGES correspond to l-mers
→ instead of vertices(Hamiltonian) - construct of graph
- vertices ar all (l-1)-mers
- arc from p to q if spectrum(s, l) contains l-mer with prefix p and suffix q
→ Then, one-to-one correspondence between trails visiting each edge exactly one in graph.
< Example >
Conclusion
- Use Hamiltonian path when we solve SBH problem
- Not an efficient algorithm
- Not practically useful for large overlap graphs
