Modeling
Data Load
import datetime
import sys
import os
import re
import io
import json
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import ta
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import KFold
from sklearn.ensemble import RandomForestClassifier, BaggingClassifier
from sklearn.metrics import accuracy_score, precision_score, recall_score, confusion_matrix, f1_score, roc_auc_score, roc_curve
sys.path.append('/aiffel/aiffel/fnguide/data/')
from libs.mlutil.pkfold import PKFold
DATA_PATH = '/aiffel/aiffel/fnguide/data/'
data_file_name = os.path.join(DATA_PATH, 'sub_upbit_eth_min_feature_labels.pkl')
df_data = pd.read_pickle(data_file_name)
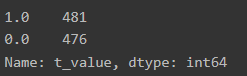
df_data['t_value'] = df_data['t_value'].apply(lambda x: x if x == 1 else 0)
df_data['t_value'].value_counts()
Scaling
X, y = df_data.iloc[:, 5:-1], df_data.iloc[:, -1]
sc = StandardScaler()
X_sc = sc.fit_transform(X)
Train, Test 분리
train_x, test_x, train_y, test_y = X_sc[:n_train, :], X_sc[-n_test:, :], y.iloc[:n_train], y.iloc[-n_test:]
train_x = pd.DataFrame(train_x, index=train_y.index, columns=X.columns)
train_y = pd.Series(train_y, index=train_y.index)
test_x = pd.DataFrame(test_x, index=test_y.index, columns=X.columns)
test_y = pd.Series(test_y, index=test_y.index)
# 학습 시간 단축을 위해 여기선 편의상 1000개의 데이터만 가져옵니다.
train_x = train_x[:1000]
train_y = train_y[:1000]
PK-FOLD 사용
학습데이터와 검증데이터를 나눌 때 두 시계열 사이의 연관성을 최대한 배제하기 위한 방법
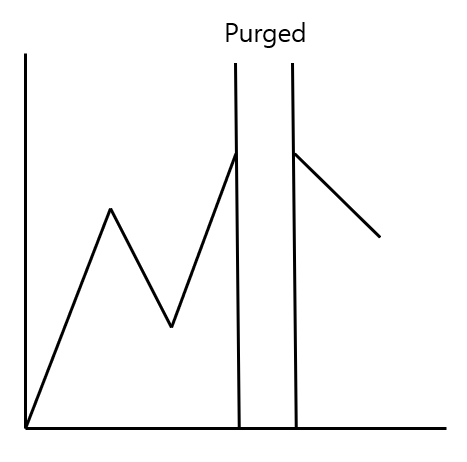
자기 상관을 낮춤
gs_rfc_best.fit(train_x, train_y)
n_cv = 4
t1 = pd.Series(train_y.index.values, index=train_y.index)
# purged K-Fold
cv = PKFold(n_cv, t1, 0)
GridSearch CV
# 최대 20분정도가 소요됩니다. 시간이 부족하다면 파라미터를 조절하여 진행하세요.
bc_params = {'n_estimators': [5, 10, 20],
'max_features': [0.5, 0.7],
'base_estimator__max_depth': [3,5,10,20],
'base_estimator__max_features': [None, 'auto'],
'base_estimator__min_samples_leaf': [3, 5, 10],
'bootstrap_features': [False, True]
}
rfc = RandomForestClassifier(class_weight='balanced')
bag_rfc = BaggingClassifier(rfc)
gs_rfc = GridSearchCV(bag_rfc, bc_params, cv=cv, n_jobs=-1, verbose=1)
gs_rfc.fit(train_x, train_y)
gs_rfc_best = gs_rfc.best_estimator_
Modeling
gs_rfc_best.fit(train_x, train_y)
pred_y = gs_rfc_best.predict(test_x)
prob_y = gs_rfc_best.predict_proba(test_x)
confusion = confusion_matrix(test_y, pred_y)
accuracy = accuracy_score(test_y, pred_y)
precision = precision_score(test_y, pred_y)
recall = recall_score(test_y, pred_y)
print('=======================================================')
print(f'정확도:{accuracy}, 정밀도:{precision}, 재현율:{recall}')

fpr, tpr, thresholds = roc_curve(test_y, pred_y)
auc = roc_auc_score(test_y, pred_y)
plt.plot(fpr, tpr, linewidth=2)
plt.plot([0, 1], [0, 1], 'k--') # dashed diagonal
plt.xlabel('fpr')
plt.ylabel('tpr')
print(f'auc:{auc}')
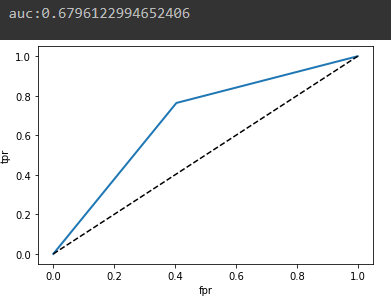
TPR : True를 True라고 정상적으로 예측
FPR : True를 False라고 잘못된 예측
ROC Curve가 좌상단으로 휠 경우 이진 분류를 잘한다는 뜻




