import pandas as pd
import matplotlib.pyplot as plt
import os
import numpy as np
import seaborn as sns
from tensorflow import keras
from tensorflow.keras import layers
from sklearn.metrics import mean_absolute_error, mean_squared_error
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler Data Summary
#Paths of data files
all_files =[]
for root, dirs, files in os.walk('/kaggle/input/skab-anomaly-detection/SKAB-master/data/'):
for file in files:
if file.endswith('.csv'):
all_files.append(os.path.join(root,file))
#Data with outliers included
list_of_df = [pd.read_csv(file,
sep = ';',
index_col = 'datetime',
) for file in all_files if 'anomaly-free' not in file]
#Anomaly free data
anomaly_free_df = pd.read_csv([file for file in all_files if 'anomaly-free' in file][0],
sep = ';',
index_col = 'datetime')print('이상치 포함된 DataFrame {}개'.format(len(list_of_df)))
for i in range(len(list_of_df)) : print(list_of_df[i].columns, list_of_df[i].shape)
print('Number of change points :',sum([len(df[df.changepoint == 1.0])for df in list_of_df]) )
print('Number of outliers :',sum([len(df[df.anomaly == 1.0])for df in list_of_df]) )
print('\n이상치 없는 데이터')
print(anomaly_free_df)
df = list_of_df[1] #두번째 데이터로 실험진행!
#시각화
df.plot(figsize = (12,6))
plt.xlabel('Time')
plt.ylabel('Value')
plt.title('Signals Visualization')
plt.show()
#데이터 분포 확인
df['anomaly'].plot(figsize = (12,3))
df['changepoint'].plot()
plt.legend()
plt.title('Distribution of Outliers ')
plt.show()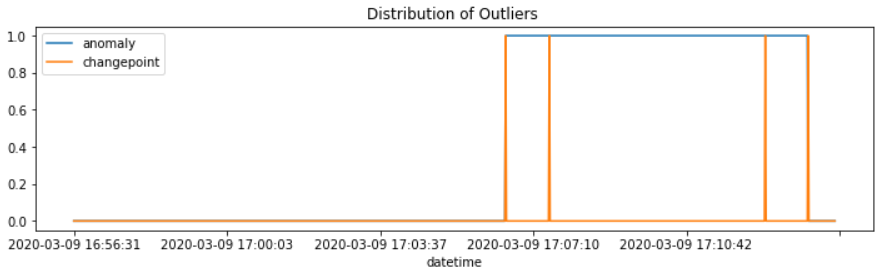
Model Pipeline and Preprocessing
# Random Seed
def Random(seed_value):
import os
os.environ['PYTHONHASHSEED'] = str(seed_value)
import random
random.seed(seed_value)
np.random.seed(seed_value)
import tensorflow as tf
tf.random.set_seed(seed_value)
#Convolutional AutoEncoder
def convae(data):
model = keras.Sequential([
layers.Input(shape = (data.shape[1], data.shape[2])),
layers.Conv1D(filters = 32, kernel_size = 7, padding = 'same', strides = 2, activation = 'relu' ),
layers.Conv1D(filters = 16, kernel_size = 7, padding = 'same', strides = 2, activation = 'relu' ),
layers.Conv1DTranspose(filters = 16, kernel_size = 7, padding = 'same', strides = 2, activation = 'relu' ),
layers.Conv1DTranspose(filters = 32, kernel_size = 7, padding = 'same', strides = 2, activation = 'relu' ),
layers.Conv1DTranspose(filters = 8, kernel_size = 7, padding = 'same'), # shape 일치시키기 위한 layer(오토인코더 구조)
])
model.compile(optimizer = keras.optimizers.Adam(learning_rate = 0.001), loss = 'mse')
model.summary()
# Training
history = model.fit(
data,
data, # data X instead of Y because it's unsupervised learning
epochs = 100,
batch_size = 32,
verbose = 0,
# early stopping 자동 학습 중단. patience 5번 loss 상승시 중단
callbacks = [
keras.callbacks.EarlyStopping(monitor = 'val_loss', patience = 5, verbose = 0)
]
)
return history, model
#Preprocessing
X_train = df[:400].drop(columns = ['anomaly','changepoint'], axis = 1) ##0~400번까지 모두 정상데이터이므로 학습데이터로 선정
StSc = StandardScaler()
X_train_scale = StSc.fit(X_train) ## Standardization(표준) : train data의 평균, 분산분포를 저장. 추후에 test data에도 똑같이 적용필요
X_train_scale = StSc.transform(X_train)
# sequence를 고려한 전처리
def create_sequences(data, time_steps = 60):
out = []
for i in range(len(data) - time_steps + 1):
out.append(data[i : (i + time_steps)])
return np.stack(out)
X_train_seq = create_sequences(X_train_scale,60) #400의 정상데이터를 60개의 묶음으로 1씩 shift 하며 묶음 -> 341개의 sequential data
X_train_seq.shape (341, 60, 8)
history, model = convae(X_train_seq)Model: "sequential_14"
Layer (type) Output Shape Param#
=================================================================
conv1d_28 (Conv1D) (None, 30, 32) 1824
_________________________________________________________________
conv1d_29 (Conv1D) (None, 15, 16) 3600
_________________________________________________________________
conv1d_transpose_42 (Conv1DT (None, 30, 16) 1808
_________________________________________________________________
conv1d_transpose_43 (Conv1DT (None, 60, 32) 3616
_________________________________________________________________
conv1d_transpose_44 (Conv1DT (None, 60, 8) 1800
=================================================================
Total params: 12,648
Trainable params: 12,648
Non-trainable params: 0
_________________________________________________________________
Epoch 1/100
11/11 - 1s - loss: 0.9466
Epoch 2/100
11/11 - 0s - loss: 0.8634
Epoch 3/100
11/11 - 0s - loss: 0.7703
Epoch 4/100
11/11 - 0s - loss: 0.6877
Epoch 5/100
11/11 - 0s - loss: 0.6260
Epoch 6/100
11/11 - 0s - loss: 0.5748
Epoch 7/100
11/11 - 0s - loss: 0.5278
Epoch 8/100
11/11 - 0s - loss: 0.4856
Epoch 9/100
11/11 - 0s - loss: 0.4477
Epoch 10/100
11/11 - 0s - loss: 0.4177
Epoch 11/100
11/11 - 0s - loss: 0.3936
Epoch 12/100
11/11 - 0s - loss: 0.3728
Epoch 13/100
11/11 - 0s - loss: 0.3549
Epoch 14/100
11/11 - 0s - loss: 0.3396
Epoch 15/100
11/11 - 0s - loss: 0.3256
Epoch 16/100
11/11 - 0s - loss: 0.3132
Epoch 17/100
11/11 - 0s - loss: 0.3011
Epoch 18/100
11/11 - 0s - loss: 0.2899
Epoch 19/100
11/11 - 0s - loss: 0.2791
Epoch 20/100
11/11 - 0s - loss: 0.2700
Epoch 21/100
11/11 - 0s - loss: 0.2618
Epoch 22/100
11/11 - 0s - loss: 0.2543
Epoch 23/100
11/11 - 0s - loss: 0.2466
Epoch 24/100
11/11 - 0s - loss: 0.2400
Epoch 25/100
11/11 - 0s - loss: 0.2346
Epoch 26/100
11/11 - 0s - loss: 0.2292
Epoch 27/100
11/11 - 0s - loss: 0.2240
Epoch 28/100
11/11 - 0s - loss: 0.2202
Epoch 29/100
11/11 - 0s - loss: 0.2161
Epoch 30/100
11/11 - 0s - loss: 0.2113
Epoch 31/100
11/11 - 0s - loss: 0.2076
Epoch 32/100
11/11 - 0s - loss: 0.2051
Epoch 33/100
11/11 - 0s - loss: 0.2018
Epoch 34/100
11/11 - 0s - loss: 0.1980
Epoch 35/100
11/11 - 0s - loss: 0.1947
Epoch 36/100
11/11 - 0s - loss: 0.1919
Epoch 37/100
11/11 - 0s - loss: 0.1894
Epoch 38/100
11/11 - 0s - loss: 0.1861
Epoch 39/100
11/11 - 0s - loss: 0.1835
Epoch 40/100
11/11 - 0s - loss: 0.1811
Epoch 41/100
11/11 - 0s - loss: 0.1793
Epoch 42/100
11/11 - 0s - loss: 0.1765
Epoch 43/100
11/11 - 0s - loss: 0.1742
Epoch 44/100
11/11 - 0s - loss: 0.1724
Epoch 45/100
11/11 - 0s - loss: 0.1702
Epoch 46/100
11/11 - 0s - loss: 0.1680
Epoch 47/100
11/11 - 0s - loss: 0.1659
Epoch 48/100
11/11 - 0s - loss: 0.1646
Epoch 49/100
11/11 - 0s - loss: 0.1626
Epoch 50/100
11/11 - 0s - loss: 0.1612
Epoch 51/100
11/11 - 0s - loss: 0.1599
Epoch 52/100
11/11 - 0s - loss: 0.1583
Epoch 53/100
11/11 - 0s - loss: 0.1567
Epoch 54/100
11/11 - 0s - loss: 0.1552
Epoch 55/100
11/11 - 0s - loss: 0.1533
Epoch 56/100
11/11 - 0s - loss: 0.1524
Epoch 57/100
11/11 - 0s - loss: 0.1516
Epoch 58/100
11/11 - 0s - loss: 0.1496
Epoch 59/100
11/11 - 0s - loss: 0.1485
Epoch 60/100
11/11 - 0s - loss: 0.1472
Epoch 61/100
11/11 - 0s - loss: 0.1465
Epoch 62/100
11/11 - 0s - loss: 0.1445
Epoch 63/100
11/11 - 0s - loss: 0.1439
Epoch 64/100
11/11 - 0s - loss: 0.1422
Epoch 65/100
11/11 - 0s - loss: 0.1420
Epoch 66/100
11/11 - 0s - loss: 0.1407
Epoch 67/100
11/11 - 0s - loss: 0.1402
Epoch 68/100
11/11 - 0s - loss: 0.1391
Epoch 69/100
11/11 - 0s - loss: 0.1386
Epoch 70/100
11/11 - 0s - loss: 0.1367
Epoch 71/100
11/11 - 0s - loss: 0.1357
Epoch 72/100
11/11 - 0s - loss: 0.1346
Epoch 73/100
11/11 - 0s - loss: 0.1338
Epoch 74/100
11/11 - 0s - loss: 0.1332
Epoch 75/100
11/11 - 0s - loss: 0.1323
Epoch 76/100
11/11 - 0s - loss: 0.1313
Epoch 77/100
11/11 - 0s - loss: 0.1306
Epoch 78/100
11/11 - 0s - loss: 0.1304
Epoch 79/100
11/11 - 0s - loss: 0.1291
Epoch 80/100
11/11 - 0s - loss: 0.1292
Epoch 81/100
11/11 - 0s - loss: 0.1272
Epoch 82/100
11/11 - 0s - loss: 0.1262
Epoch 83/100
11/11 - 0s - loss: 0.1258
Epoch 84/100
11/11 - 0s - loss: 0.1254
Epoch 85/100
11/11 - 0s - loss: 0.1243
Epoch 86/100
11/11 - 0s - loss: 0.1237
Epoch 87/100
11/11 - 0s - loss: 0.1233
Epoch 88/100
11/11 - 0s - loss: 0.1227
Epoch 89/100
11/11 - 0s - loss: 0.1218
Epoch 90/100
11/11 - 0s - loss: 0.1212
Epoch 91/100
11/11 - 0s - loss: 0.1205
Epoch 92/100
11/11 - 0s - loss: 0.1205
Epoch 93/100
11/11 - 0s - loss: 0.1196
Epoch 94/100
11/11 - 0s - loss: 0.1189
Epoch 95/100
11/11 - 0s - loss: 0.1179
Epoch 96/100
11/11 - 0s - loss: 0.1170
Epoch 97/100
11/11 - 0s - loss: 0.1164
Epoch 98/100
11/11 - 0s - loss: 0.1159
Epoch 99/100
11/11 - 0s - loss: 0.1157
Epoch 100/100
11/11 - 0s - loss: 0.1149Anomaly Scoring & Result
residuals = pd.Series(np.sum(np.mean(np.abs(X_train_seq - model.predict(X_train_seq)), axis = 1), axis = 1)) # input data와 reconstructed input의 차이값
ucl = residuals.quantile(0.999) # 차이값의 전체 분포에서 상위 0.01% 지점을 upper control limit으로 지정
plt.plot(residuals) ## value gap between original input and reconstructed input for anomaly-free data
plt.show()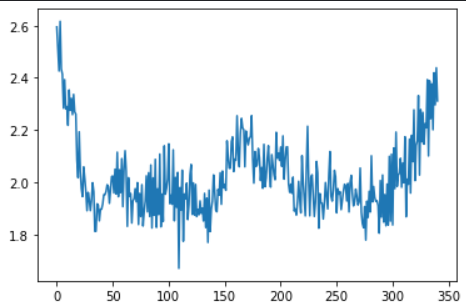
X_test_sc = StSc.transform(df.drop(columns = ['anomaly','changepoint'])) #400번째 이후의 이상치 데이터 모두 포함하여 test data 지정
X_test_seq = create_sequences(X_test_sc)
test_residuals = pd.Series(np.sum(np.mean(np.abs(X_test_seq - model.predict(X_test_seq)), axis = 1), axis = 1))
plt.figure(figsize = (15,5))
plt.subplot(1,2,1)
plt.plot(test_residuals)
plt.axhline(y = ucl * 1.5 , xmin = 0, xmax=len(X_train_seq)) # upper control limit 의 1.5배를 Threshold 로 설정
plt.title('Reconstructed Residual Error')
plt.subplot(1,2,2)
plt.plot(df['anomaly'].values)
plt.title('Actual Label')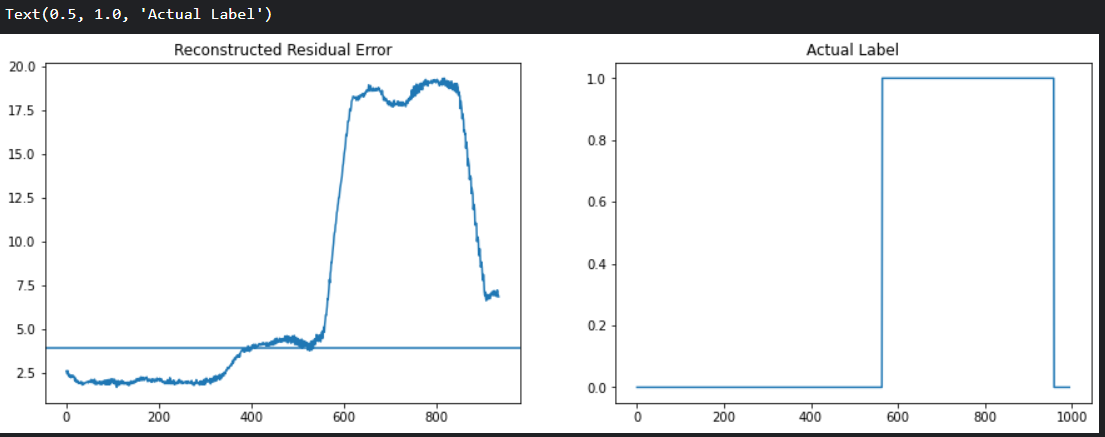
# 비교를 위해 label값을 time 단위로 변경
N_STEPS = 60
anomalous_data = test_residuals > ucl * 1.5
anomalous_data_indices = []
for data_idx in range(N_STEPS - 1, len(X_test_seq) - N_STEPS + 1):
if np.all(anomalous_data[data_idx - N_STEPS + 1 : data_idx]):
anomalous_data_indices.append(data_idx)
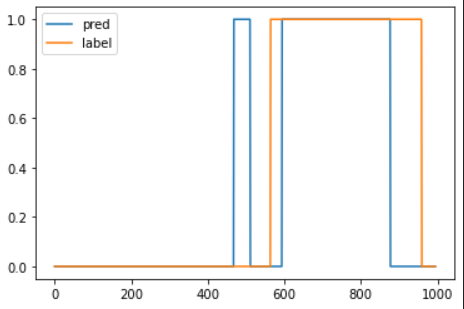
prediction = pd.Series(data = 0, index = df.index)
prediction.iloc[anomalous_data_indices] = 1
plt.plot(prediction.values, label = 'pred')
plt.plot(df['anomaly'].values, label = 'label')
plt.legend()
plt.show()