Constraint-based Algorithm_pc

Note : PC Algorithm
Pseudo Code
[Assumption]
- No hidden confounder Assumption
- Markov Causal Assumption
- Faithfulness Assumption
Problem : Given a set IND of d-separations, determine the DAG pattern faithful
to IND if there is one
Input : a set V of nodes and a set IND of d-separations among subsets of the
nodes
Outputs : If IND admits a faithful DAG representation, the DAG pattern gp
containing the d-separations in this set
# Warning : the notations in this code are not in the original code. I wrote it.
# So, the notations are possibly wrong
def PC(V, IND) -> Graph G:
S[(x,y)] <- the set Sxy that render X and Y are conditional independent
# step 0 and step 1
# Lemma 1, Verma and Pearl, 1991, On the Equivalence of Causal Models
# X and Y are adjacent
# <-> X and Y are not d-separated by any subset of PA_X or PA_Y
# => X and Y are not adjacent
# <-> X and Y are d-separated by some subset of PA_X or PA_Y
# therefore, we do not have to check d-separation for every possible subset S,
# only have to do for subsets of PA_X or PA_Y
G <- the complete undirected graph over V
i = 0
while |ADJ_X| > i for any X in V:
for each X in V:
for each Y in ADJ_X:
if there is a set S_ <= ADJ_X/{Y} such that |S_| = i and I({X},{Y}|S_) in IND:
S[(X,Y)] = S_
remove the link X-Y from G
i += 1
~~~~~~~~~
SAME WITH STEP 2, 3, 4, 5 in Basic algorithm
~~~~~~~~~
return G❗ The original code is C-style, so I modify the code to be similar to Python style
❓ What is 'Basic algorithm'? ⇒ See Constraint-based Algorithm_basic
Even in the worst-case, PC algorithm has the same time complexity as Basic algorithm and it becomes reasonably efficient if the graph becomes sparse. [3]
Soundness & Completeness of PC algorithm
The result of step 0 & 1 in PC algorithm is same with the one of step 0 & 1 in Basic algorithm.[4] The other steps are equal with Basic algorithm. Therefore, the DAG pattern generated by PC algorithm is sound and complete.
Implementation
We only need to change step 0 & 1.
STEP 0 & 1 : Find Skeleton by relation of adjacency and d-separation ( = p-independence in practice)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from collections import deque
from itertools import combinations, chain
from collections import defaultdictdef identify_skeleton_from_full_link_graph(self, data, test_kwarg):
self.ptn = pattern()
self.ptn.add_vertex(list(data.columns))
self.p_independence_set = defaultdict(set)
self.p_independence_set = defaultdict(lambda: defaultdict(set))
# Lemma 1, Verma and Pearl, 1991, On the Equivalence of Causal Models
# X and Y are adjacent
# <-> X and Y are not d-separated by any subset of PA_X or PA_Y
# => X and Y are not adjacent
# <-> X and Y are d-separated by some subset of PA_X or PA_Y
# therefore, we do not have to check d-separation for every possible subset S,
# only have to do for subsets of PA_X or PA_Y
self.ptn.full_link()
adj = {x : self.ptn.adjacent(x) for x in self.ptn.vertex}
i = 0
while any(i < len(adj[x]) for x in adj.keys()):
for x in self.ptn.vertex:
adj_x = adj[x]
for y in adj_x:
adj_x_not_y = list(adj_x - {y})
power_set = combinations(adj_x_not_y, i)
for subset in power_set:
if self.test(data, {x}, {y}, set(subset), **test_kwarg):
self.p_independence_set[x][y]
self.p_independence_set[y][x] = self.p_independence_set[x][y]
self.p_independence_set[x][y].add(subset)
self.ptn.remove_links([(x, y)])
break
adj[x] = self.ptn.adjacent(x)
i += 1class pc
pip install cdpi -qfrom cdpi import pattern
from cdpi.causal_discovery.test import get_test
from cdpi.causal_discovery.util import (identify_skeleton_by_ind,
identify_v_structure_with_adjacency_orient_faithfulness,
identify_meeks_rule_2,
identify_meeks_rule_3,
identify_meeks_rule_4 )class pc:
def __init__(self):
self.ptn = pattern()
def identify(self, data:pd.DataFrame = None, test:str = None, ind:dict = None, vertex = None, **test_kwarg) -> pattern:
# STEP 0 ~ 1 : Find skeleton
if ind is not None:
self.identify_skeleton_by_ind(ind, vertex)
elif data is not None and test is not None:
self.test = get_test(test)
self.identify_skeleton_from_full_link_graph(data, test_kwarg = test_kwarg)
else:
print("pc.identify : both ind and (data, test) are None!")
# # STEP 2 : Find v-structure
self.identify_v_structure_with_adjacency_orient_faithfulness()
# STEP 3~5 : use Meek's rules
cnt = True
while cnt:
cnt2 = self.identify_meeks_rule_2()
cnt3 = self.identify_meeks_rule_3()
cnt4 = self.identify_meeks_rule_4()
cnt = cnt2 or cnt3 or cnt4 # Check there are vertexs which could be changed
return self.ptn
def draw(self):
self.ptn.draw()
def draw_by_pos(self, pos):
self.ptn.draw_by_pos(pos)
pc.identify_skeleton_from_full_link_graph = identify_skeleton_from_full_link_graph
pc.identify_v_structure_with_adjacency_orient_faithfulness = identify_v_structure_with_adjacency_orient_faithfulness
pc.identify_meeks_rule_2 = identify_meeks_rule_2
pc.identify_meeks_rule_3 = identify_meeks_rule_3
pc.identify_meeks_rule_4 = identify_meeks_rule_4
pc.identify_skeleton_by_ind = identify_skeleton_by_indExample
np.random.seed(seed=2022)Discrete
import scipy.stats as stats
size = 500
data = pd.DataFrame()
data['A'] = stats.bernoulli(p = 0.5).rvs(size = size)
data['B'] = stats.bernoulli(p = 0.5).rvs(size = size)
data['C'] = stats.bernoulli(p = 0.5).rvs(size = size)
data['D'] = data['A'] + data['B'] + data['C'] + stats.bernoulli(p = 0.5).rvs(size = size)
data['E'] = data['A'] + data['B'] + stats.bernoulli(p = 0.5).rvs(size = size)
pc_al = pc()
pc_al.identify(data, test = 'conditional_mutual_information_test')
pc_al.draw()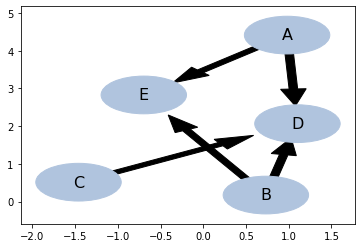
For given IND
ptn = pattern()
ptn.add_edges([
('A', 'D'),
('D', 'B'),
('B', 'E'),
('C', 'E'),
('C', 'F'),
('D', 'H'),
('E', 'I'),
('F', 'I'),
('H', 'K'),
('I', 'L'),
('J', 'L'),
('G', 'J')])
ptn.draw()
pos = ptn.pos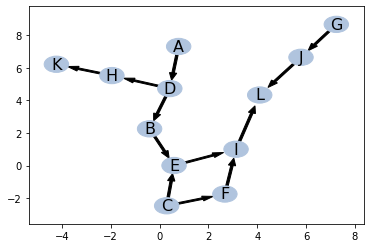
ind = ptn.get_all_d_separation()pc_al = pc()
pc_al.identify(ind = ind)
pc_al.draw_by_pos(pos)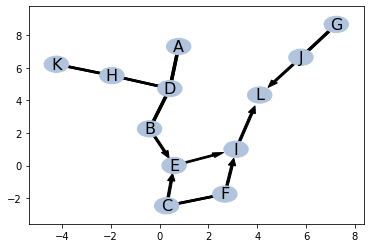
Reference
[3] Metropolitan. (2004). Learning Bayesian Networks. chapter 10. pp.546.
[4] Verma, T., Pearl, J. (1990). On the Equivalence of Causal Models. Appears in Proceedings of the Sixth Conference on Uncertainty in Artificial Intelligence (UAI1990).