https://www.kaggle.com/datasets/iarunava/cell-images-for-detecting-malaria
Malaria cell
- 주제 : 말라리아 혈액도말 이미지 분류 실습
- 목적 : TF공식 문서의 이미지 분류를 다른 이미지를 사용해서 응용
- 이미지 데이터 불러오기 'wget'을 사용하면 온라인 url에 있는 파일을 불러올 수 있다. 논문(혈액도말 이미지로 말라리아 감염여부를 판단하는 논문)에 사용한 데이터셋을 불러옴
- plt.imread와 CV2(OpenCV)의 imread를 통해 array형태로 데이터를 불러와서 시각화를 해서 감염된 이미지와 아닌 이미지를 비교
- TF.kera의 전처리 도구를 사용해서 train, valid set을 나눠줌 -> 레이블 값을 폴더명으로 생성해주게 됨
라이브러리 로드
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# layers 에서는 Conv2D, MaxPool2D, Dropout, Flatten, Dense 를 불러옵니다.
# callbacks 에서는 EarlyStopping 을 불러옵니다.
import tensorflow as tf
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Conv2D, MaxPool2D, Dropout, Flatten, Dense
from tensorflow.keras.callbacks import EarlyStopping이미지 다운로드 & 압축 해제
!wget https://data.lhncbc.nlm.nih.gov/public/Malaria/cell_images.zip
!unzip cell_images.zipimport os
for root, dirs, files in os.walk("./cell_images/"):
print(root, dirs, len(files))
>>>>
./cell_images/ ['Parasitized', 'Uninfected'] 0
./cell_images/Parasitized [] 13780
./cell_images/Uninfected [] 13780일부 이미지 미리보기
import glob
upics = glob.glob('./cell_images/Uninfected/*.png')
apics = glob.glob('./cell_images/Parasitized/*.png')
len(upics), upics[0], len(apics), apics[0]
>>>>
(13779,
'./cell_images/Uninfected/C174P135NThinF_IMG_20151127_135311_cell_143.png',
13779,
'./cell_images/Parasitized/C168P129ThinF_IMG_20151118_154126_cell_159.png')# upics
upics_0 = upics[0]
upics_0_img = plt.imread(upics_0)
plt.imshow(upics_0_img) 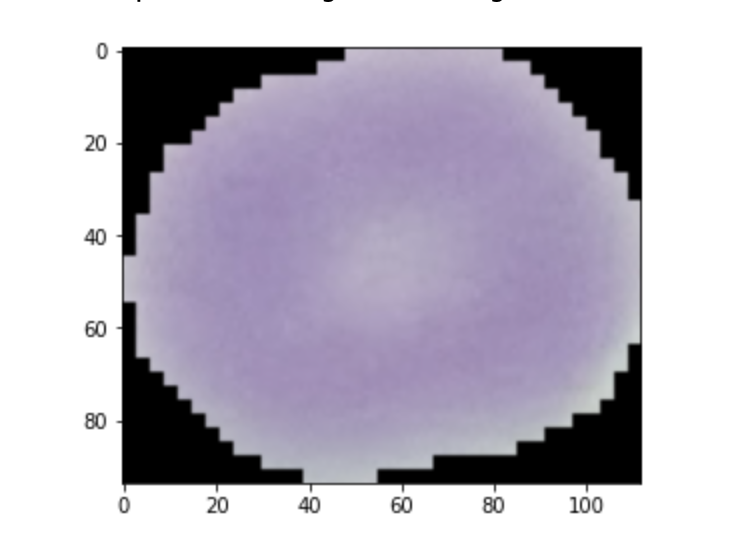
# apics
apics_0 = apics[0]
apics_0_img = plt.imread(apics_0)
plt.imshow(apics_0_img) 
cv2로 시각화
Uninfected
import cv2
plt.figure(figsize=(8, 8))
labels = 'Uninfected'
for i images in enumerate(upics[:9]):
ax = plt.subplot(3, 3, i+1)
img = cv2.imread(images)
plt.imshow(img)
plt.title(f'{labels} {img.shape}')
plt.axis('off')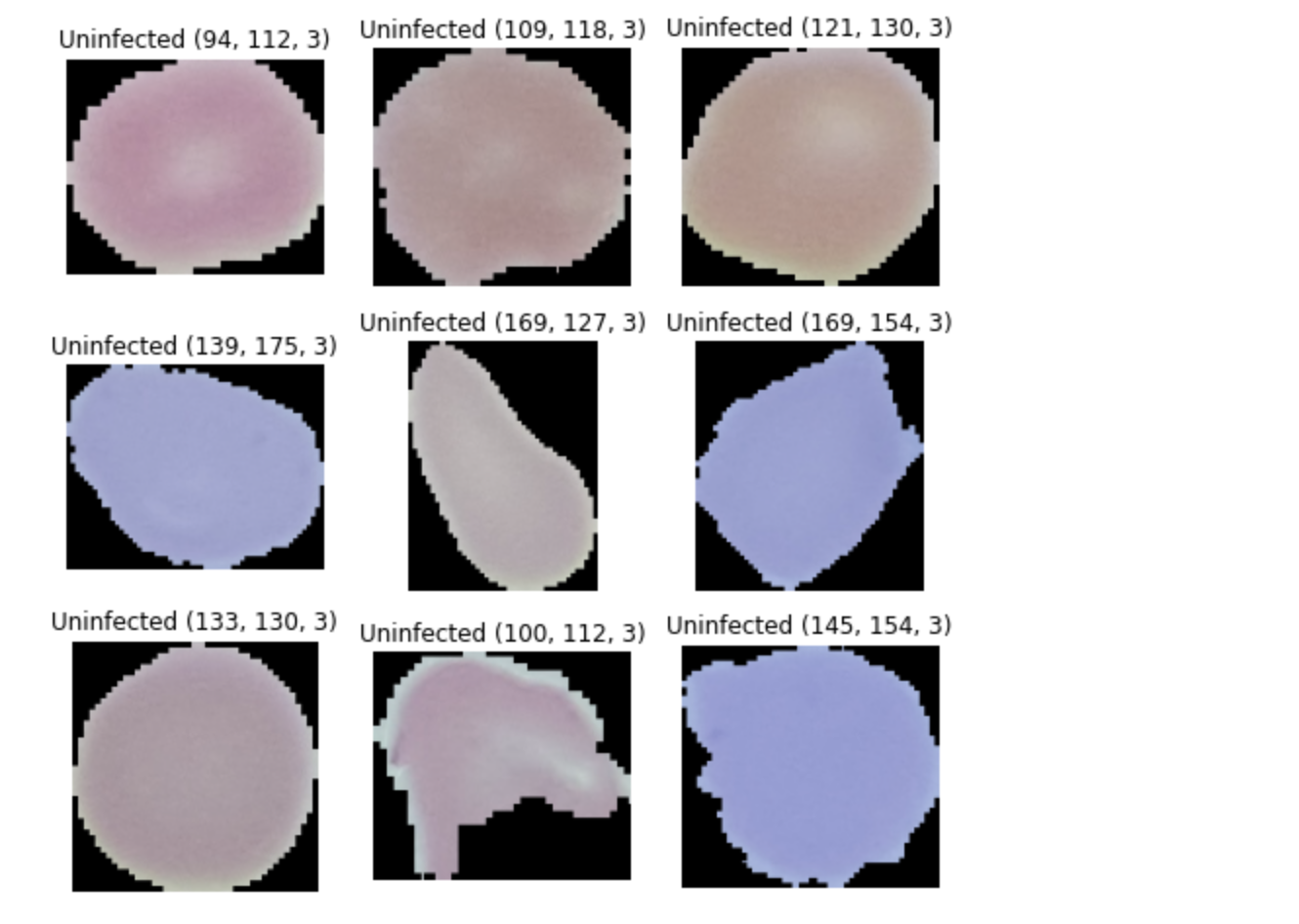
Infected
plt.figure(figsize=(8, 8))
labels = "Infected"
for i, images in enumerate(apics[:9]):
ax = plt.subplot(3, 3, i + 1)
img = cv2.imread(images)
plt.imshow(img)
plt.title(f'{labels} {img.shape}')
plt.axis("off")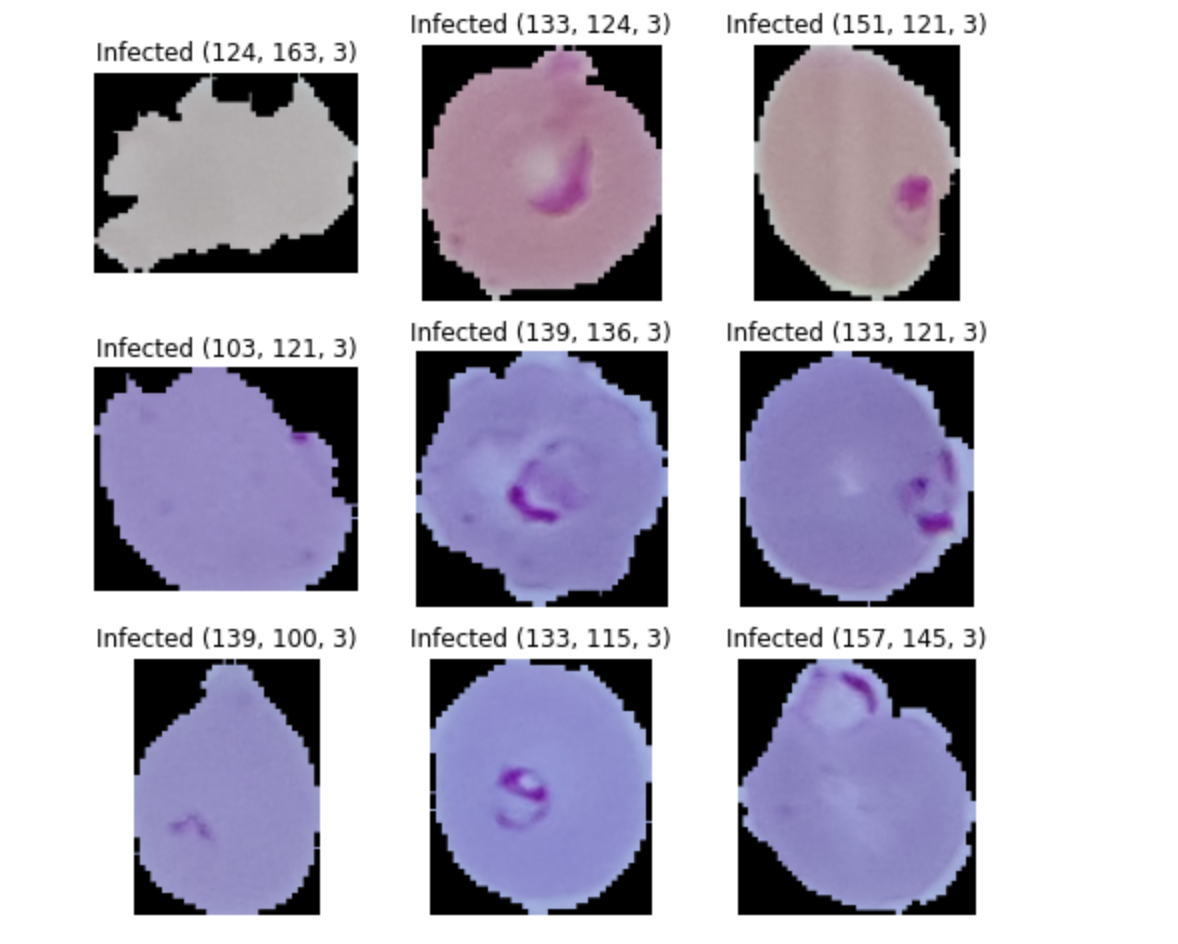
데이터셋 나누기
- 학습, 검증 세트로 나누기
- ImageDataGenerator 통해 이미지를 로드하고 전처리
from tensorflow.keras.preprocessing.image import ImageDataGenerator
#validation_split 값을 통해 학습 : 검증 비율을 8:2로 나눈다
datagen = ImageDataGenerator(rescale=1/255.0, validation_split=0.2)이미지 사이즈 설정
- 이미지의 사이즈가 불규칙하면 학습을 할 수 없기 때문에 리사이즈할 크기를 지정
- 이미지가 너무 작으면 왜곡되거나 특징을 잃어버릴 수 있다 하지만 계산량이 줄어들어 학습속도가 빠르다.
- 이미지가 너무 크면 흐려지거나 왜곡이 될 수 있으나 좀 더 자세하게 학습할 수도 있다. 하지만 계산량이 많아 학습속도가 오래걸린다.
width=16
height=16학습 세트
-
flow_from_directory 통해 이미지를 불러옵니다.
-
training 데이터셋 생성한다.
-
class_mode 에는 이진분류이기 때문에 binary 넣어준다
classmode : One of 'categorical', 'binary', 'sparse',
'input', or None.
Default : 'categorical'.
subset : Subset of data {'training' or 'validation'}
trainDatagen = datagen.flow_from_directory(directory = 'cell_images/',
target_size = (height, width),
class_mode = 'binary',
batch_size = 64,
subset='training')
>>>>
Found 22048 images belonging to 2 classes.trainDatagen.num_classes
>>>> 2
trainDatagen.classes
>>>>
array([0, 0, 0, ..., 1, 1, 1], dtype=int32)
# {'Parasitized' : 0, 'Uninfected' : 1}
# 0 : 감염, 1 : 미감염
trainDatagen.class_indices
>>>>
{'Parasitized': 0, 'Uninfected': 1}검증세트
: validation 데이터셋 생성
valDatagen = datagen.flow_from_directory(directory = 'cell_images/',
target_size =(height, width),
class_mode = 'binary',
batch_size = 64,
subset='validation')레이어 설정
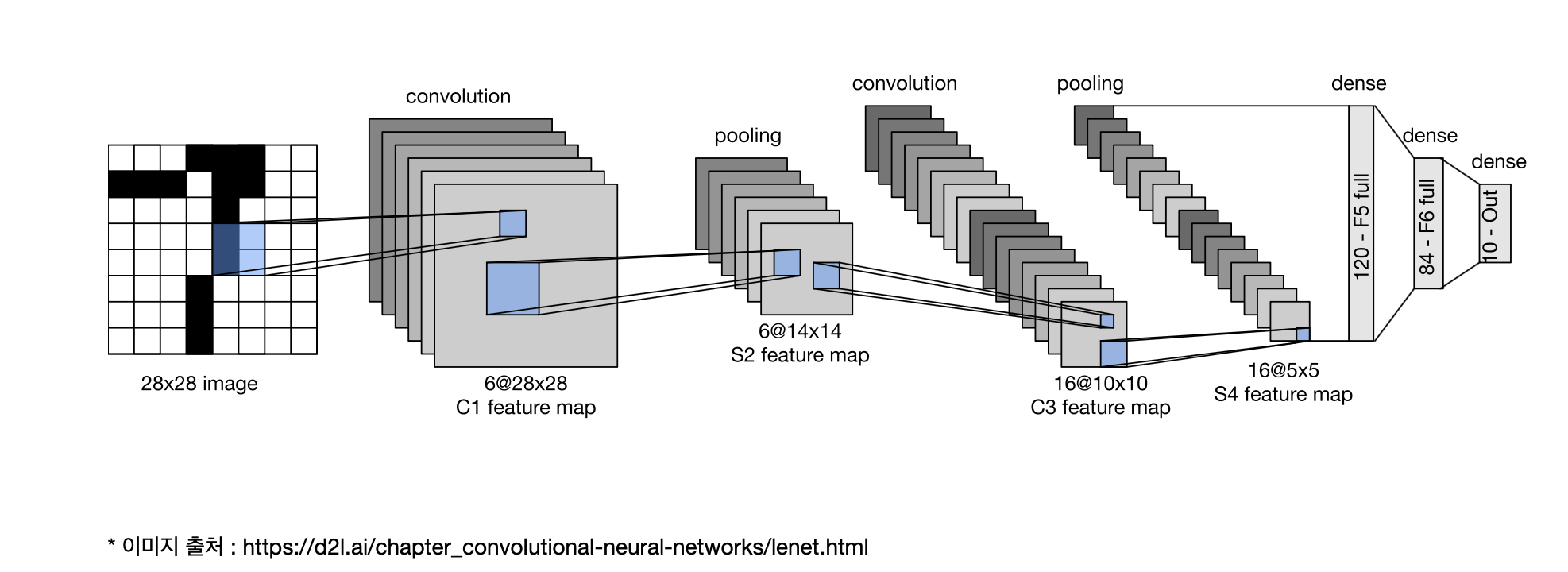
- padding : 경계 처리 방법
- 'valid' : 유효한 영역만 출력. 따라서 출력 이미지 사이즈는 입력 사이즈보다 작다.
- 'same' : 출력 이미지 사이즈가 입력 이미지 사이즈와 동일 - input_shape : 모델에서 첫 레이어에서 정의 (height, width, channels)
- activation : 활성화 함수 설정
- 'linear' : 디폴트 값, 입력 뉴런과 가중치로 계산된 결과값이 그대로 출력
- 'relu' : rectifier 함수, 은닉층에 주로 사용
- 'sigmoid' : 시그모이드 함수, 이진 분류 문제에서 출력층에 주로 사용
- 'softmax' : 소프트맥스 함수, 다중 클래스 분류 문제에서 출력층에 주로 사용 - Dense Layer :
- 밀집 연결 또는 완전 연결층이라고 불림
- 첫 번째 Dense층은 128개의 노드를 가진다
- 마지막 층은 출력층- softmax : 2개의 노드의 소프트맥스 층. 이 층은 2개의 확률을 반환하고 반환된 값의 전체 합은 1이다.
- sigmoid : 둘 중 하나를 예측할 때 1개의 출력값을 출력. 확률을 받아 임계값 기준으로 True, False로 나눈다.
- Output Layer 출력층
- 예측 값이 n개 : keras.layers.Dense(n, activation='softmax')
- 예측 값이 둘 중 하나 : keras.layers.Dense(1, activation='sigmoid')
model = Sequential()
# 입력층
model.add(Conv2D(filters=16, kernel_size=(3,3), padding='valid',
activation='relu', input_shape=(height, width, 3)))
model.add(Conv2D(filters=16, kernel_size=(3,3), padding='same',
activation='relu'))
model.add(MaxPool2D(pool_size=(2,2), strides=1))
model.add(Conv2D(filters=16, kernel_size=(3,3), padding='same',
activation='relu'))
model.add(Conv2D(filters=16, kernel_size=(3,3), padding='same',
activation='relu'))
model.add(MaxPool2D(pool_size=(2,2), strides=1))
# 20% 는 0으로 대체합니다. => 오버피팅 방지
model.add(Dropout(0.2))
# Fully-connected layer
model.add(Flatten())
model.add(Dense(units=64, activation='relu'))
model.add(Dense(units=32, activation='relu'))
# 출력층=> binary classification
model.add(Dense(1, activation='sigmoid'))
# summary
model.summary()
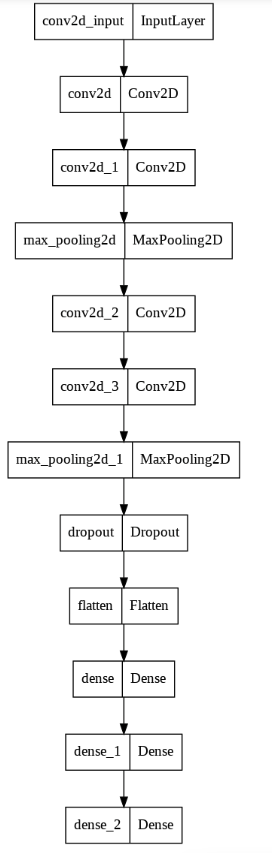
# tensorflow.keras.utils의 plot_model 을 통한 레이어 시각화
from tensorflow.keras.utils import plot_model
plot_model(model)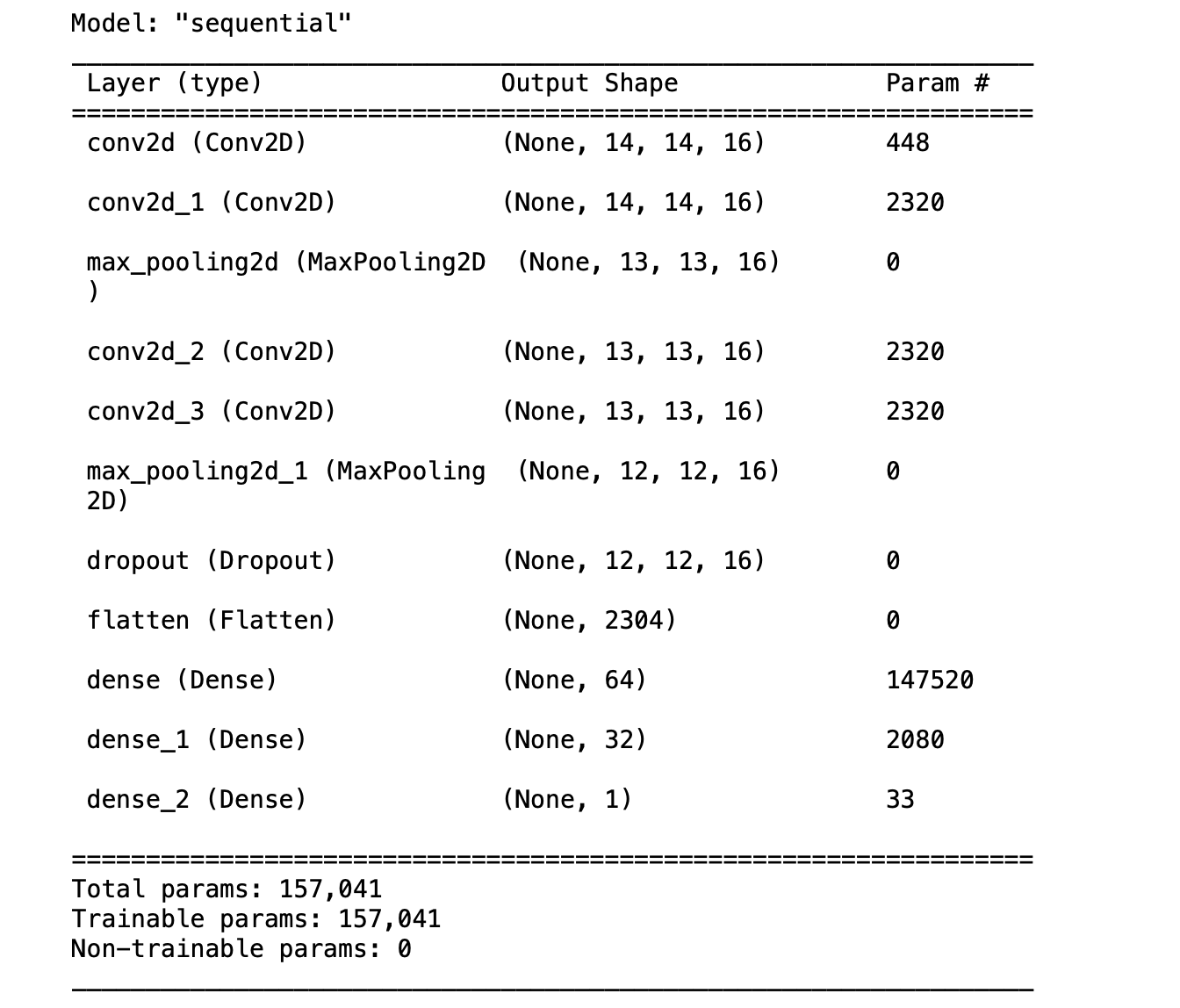
compile
model.complie(optimizer='adam',
loss='binary_crossentropy',
metrics=[accuracy'])학습 (fit)
배치(batch): 모델 학습에 한 번에 입력할 데이터셋
에폭(epoch): 모델 학습시 전체 데이터를 학습한 횟 수
스텝(step): (모델 학습의 경우) 하나의 배치를 학습한 횟 수
EarlyStopping: 성능이 더 이상 좋아지지 않으면 학습을 중지
early_stop = EarlyStopping(monitor='val_loss', patience=2)
early_stop = EarlyStopping(monitor='val_loss', patience=5)
# fit
history = model.fit(trainDatagen, epochs=1000, validation_data=valDatagen, callbacks=early_stop)
# history
df_hist = pd.DataFrame(history.history)
df_hist.tail()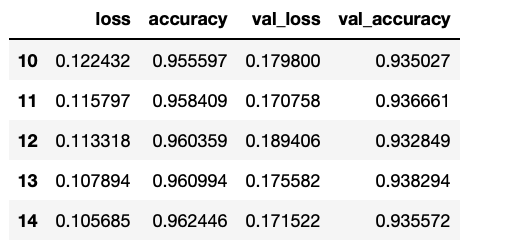
학습결과 시각화
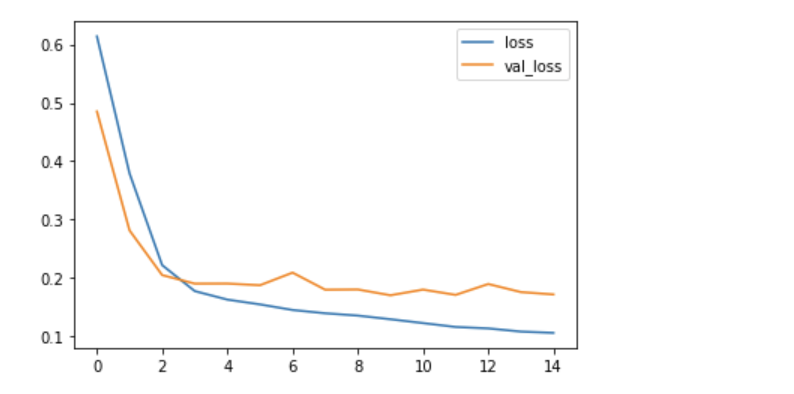
df_hist[['loss', 'val_loss']].plot()
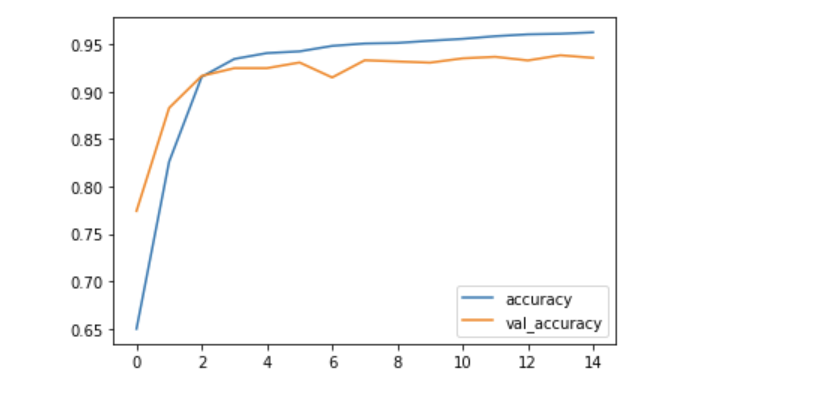
df_hist[['accuracy', 'val_accuracy']].plot()