[ML] Logistic Regression, PCA, Precision and Recall
📌 Logistic Regression
- 분류
import numpy as np
z = np.arange(-10, 10, 0.01)
g = 1 / (1+np.exp(-z))
import matplotlib.pylab as plt
%matplotlib inline
plt.plot(z, g)
plt.grid()
plt.show()
# graph detail
plt.figure(figsize=(12, 8))
ax = plt.gca() # gca: axis 설정값 변경할 수 있음
ax.plot(z, g)
# 축 설정
ax.spines['left'].set_position('zero') #
ax.spines['right'].set_color('none')
ax.spines['bottom'].set_position('center')
ax.spines['top'].set_color('none')
plt.show()
- Hypothesis 함수의 결과에 따른 분류
- Decision Boundary
Logistic Reg의 Cost Function의 그래프
h = np.arange(0.01, 1, 0.01)
C0 = -np.log(1-h)
C1 = -np.log(h)
plt.figure(figsize=(12, 8))
plt.plot(h, C0, label='y=0')
plt.plot(h, C1, label='y=1')
plt.legend()
#plt.grid()
plt.show()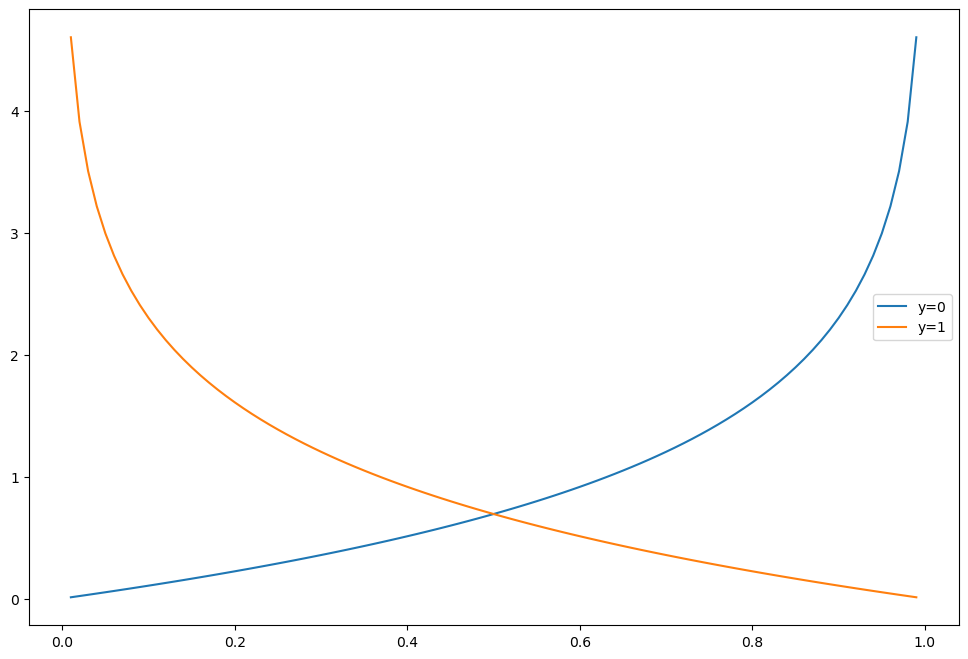
📌 실습
import pandas as pd
wine_url = 'https://raw.githubusercontent.com/PinkWink/ML_tutorial/master/dataset/wine.csv'
wine = pd.read_csv(wine_url, index_col=0)
# 맛 등급 만들어 넣기
wine['taste'] = [1. if grade>5 else 0. for grade in wine['quality']]
X = wine.drop(['taste', 'quality'], axis=1)
y = wine['taste']- 데이터 분리
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=13)- 간단 로지스틱 회귀 테스트
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
lr = LogisticRegression(solver='liblinear', random_state=13)
lr.fit(X_train, y_train)
y_pred_tr = lr.predict(X_train)
y_pred_test = lr.predict(X_test)
print('Train Acc: ', accuracy_score(y_train, y_pred_tr))
print('Test Acc: ', accuracy_score(y_test, y_pred_test))Train Acc: 0.7427361939580527
Test Acc: 0.7438461538461538
- 스케일러까지 적용해서 파이프라인 구축
- cf. DT는 스케일러의 영향 크게 받지 않음
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
estimators = [('scaler', StandardScaler()),
('clf', LogisticRegression(solver='liblinear', random_state=13))]
pipe = Pipeline(estimators)
pipe.fit(X_train, y_train)
y_pred_tr = pipe.predict(X_train)
y_pred_test = pipe.predict(X_test)
print('Train Acc: ', accuracy_score(y_train, y_pred_tr))
print('Test Acc: ', accuracy_score(y_test, y_pred_test))Train Acc: 0.7444679622859341
Test Acc: 0.7469230769230769
--> scaler에 의한 변화 쪼금 생김
- DT와 비교를 위한 작업
from sklearn.tree import DecisionTreeClassifier
wine_tree = DecisionTreeClassifier(max_depth=2, random_state=13)
wine_tree.fit(X_train, y_train)
models = {
'logistic regression': pipe, # logistic regression 은 scaler 있는 게 성능 쫌 더 좋았음
'decision tree': wine_tree # decision tree 는 scaler 영향 덜 받음
}- AUC 그려서 비교
from sklearn.metrics import roc_curve
plt.figure(figsize=(10, 8))
plt.plot([0, 1], [0, 1], label='random_guess')
for model_name, model in models.items():
pred = model.predict_proba(X_test)[:, 1] # 첫 번째 column: 0일 확률, 두 번째 column: 1일 확률이므로 두 번째 column 가져오기
fpr, tpr, thresholds = roc_curve(y_test, pred)
plt.plot(fpr, tpr, label=model_name)
plt.grid()
plt.legend()
plt.show()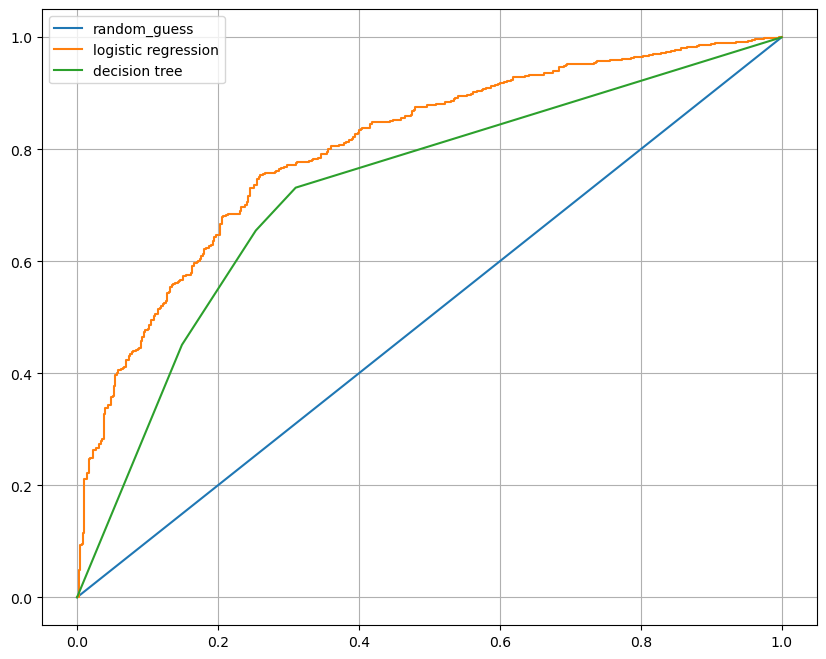
--> logistic이 성능 더 좋은 것 확인
PIMA 인디언 당뇨병 예측
PIMA_url = 'https://raw.githubusercontent.com/PinkWink/ML_tutorial/master/dataset/diabetes.csv'
PIMA = pd.read_csv(PIMA_url)
PIMA.info()>>>
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KB- float 로 데이터 변환
PIMA = PIMA.astype('float')
PIMA.info()- 상관관계 확인
import seaborn as sns
import matplotlib.pyplot as plt
plt.figure(figsize=(12, 10))
sns.heatmap(PIMA.corr(), cmap='YlGnBu') # yello green blue
plt.show()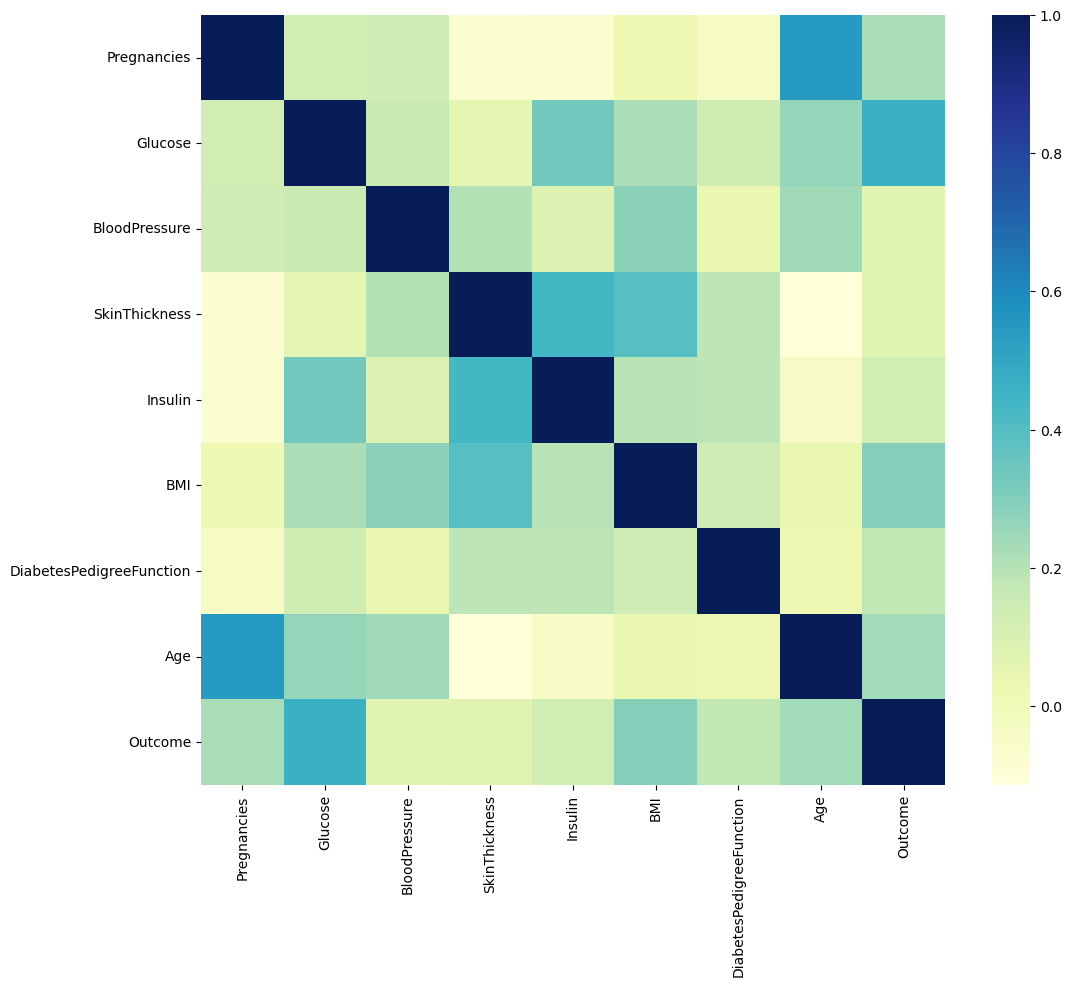
- 결측치 확인
(PIMA==0).astype(int).sum() # column별 0 갯수>>>
Pregnancies 111
Glucose 5
BloodPressure 35
SkinThickness 227
Insulin 374
BMI 11
DiabetesPedigreeFunction 0
Age 0
Outcome 500
dtype: int64- 결측치 -> 0으로 대체
zero_features = ['Glucose', 'BloodPressure', 'SkinThickness', 'BMI']
PIMA[zero_features] = PIMA[zero_features].replace(0, PIMA[zero_features].mean())(PIMA==0).astype(int).sum()>>>
Pregnancies 111
Glucose 0
BloodPressure 0
SkinThickness 0
Insulin 374
BMI 0
DiabetesPedigreeFunction 0
Age 0
Outcome 500
dtype: int64- 데이터 나누기
X = PIMA.drop(['Outcome'], axis=1)
y = PIMA['Outcome']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2,
stratify = y,
random_state=13
)- pipeline
estimators = [
('scaler', StandardScaler()),
('clf', LogisticRegression(solver='liblinear', random_state=13))
]
pipe_lr = Pipeline(estimators)
pipe_lr.fit(X_train, y_train)
pred = pipe_lr.predict(X_test)- 수치 확인
from sklearn.metrics import (accuracy_score, recall_score, precision_score, roc_auc_score, f1_score)
print('Accuracy: ', accuracy_score(y_test, pred))
print('Recall: ', recall_score(y_test, pred))
print('Precision: ', precision_score(y_test, pred))
print('AUC score: ', roc_auc_score(y_test, pred))
print('f1 score: ', f1_score(y_test, pred))Accuracy: 0.7727272727272727
Recall: 0.6111111111111112
Precision: 0.7021276595744681
AUC score: 0.7355555555555556
f1 score: 0.6534653465346535
- 다변수 방정식의 각 계수 값 확인
coeff = list(pipe_lr['clf'].coef_[0])
labels = list(X_train.columns)
coeff>>>
[0.3542658884412649,
1.2014244425037581,
-0.15840135536286706,
0.03394657712929965,
-0.1628647195398813,
0.620404521989511,
0.3666935579557874,
0.17195965447035091]- 중요 feature 그려보기
features = pd.DataFrame({'Features':labels, 'importance':coeff})
features.sort_values(by=['importance'], ascending=True, inplace=True)
features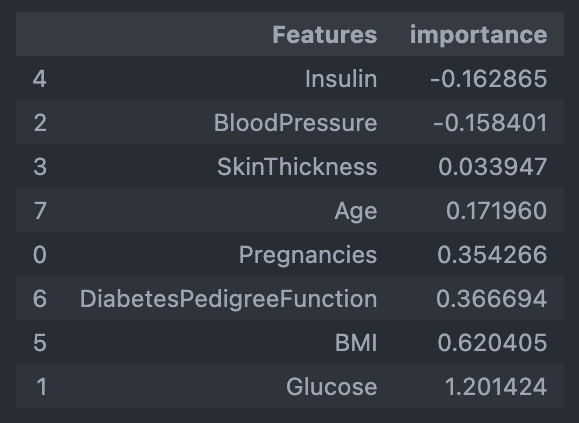
features['positive'] = features['importance'] > 0
features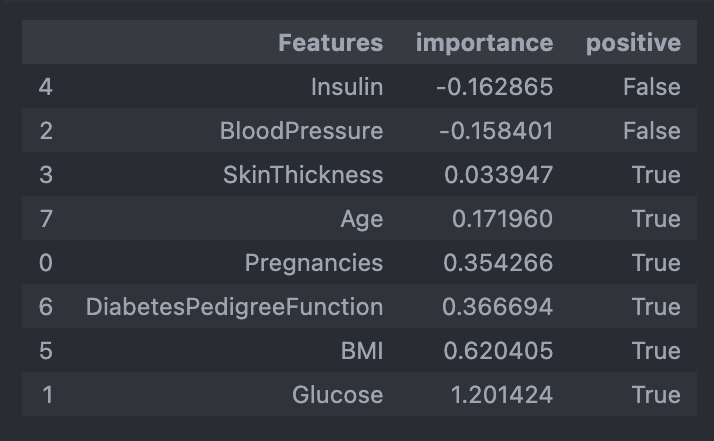
features.set_index('Features', inplace=True)
features['importance'].plot(kind='barh',
figsize=(11, 6),
color=features['positive'].map({True:'blue',
False:'red'}))
plt.xlabel('Importance')
plt.show()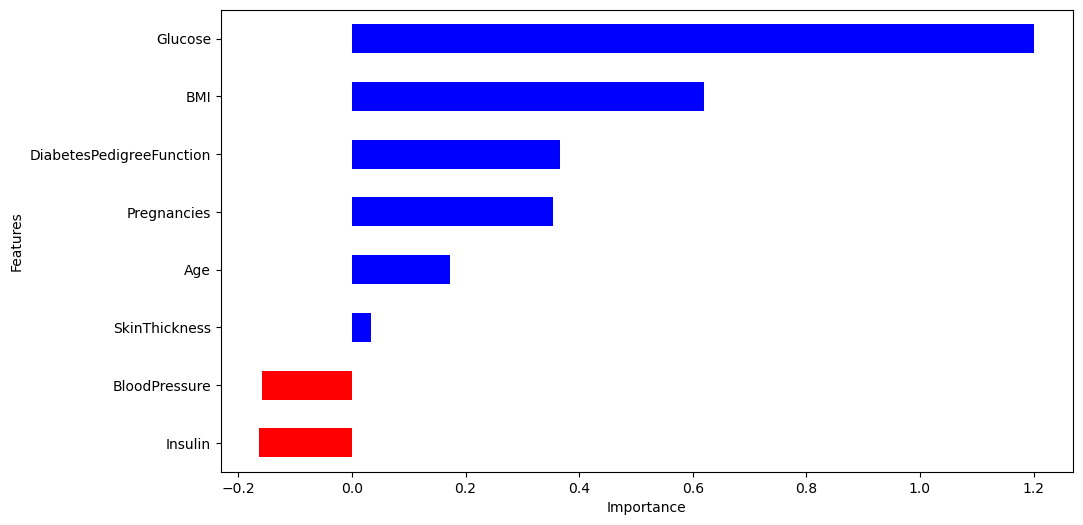
📌 PCA
데이터 만들기
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
sns.set_style('whitegrid')
rng = np.random.RandomState(13)
X = np.dot(rng.rand(2, 2), rng.randn(2, 200)).T # transpose
X.shape(200, 2)
rng.rand(2, 2) # (2, 2로 0부터 1 사이에 있는 값 4개 랜덤으로 뽑음)
rng.rand(2, 200) # 표준정규분포 따르는 랜덤 수를 (2, 200) 으로 나열
np.dot(rng.rand(2, 2), rng.randn(2, 200)).Tplt.scatter(X[:, 0], X[:,1])
plt.axis('equal') # 두 축 같은 간격으로 그리기
fit
from sklearn.decomposition import PCA
pca = PCA(n_components=2, random_state=13)
pca.fit(X)백터, 분산값
pca.components_
pca.explained_variance_ # 설명력
pca.explained_variance_ratio_ # 설명력 - 비율주성분 벡터 그리기
def draw_vector(v0, v1, ax=None):
ax = ax or plt.gca() # None 들어오면 ax=plt.gca()
arrowprops = dict(arrowstyle='->',
linewidth=2, color='black',
shrinkA=0, shrinkB=0)
ax.annotate('', v1, v0, arrowprops=arrowprops)pca.mean_ # 좌표의 중심, 데이터의 중심array([-0.03360413, -0.03527382])
plt.scatter(X[:, 0], X[:, 1], alpha=0.4)
for length, vector in zip(pca.explained_variance_, pca.components_):
v = vector * 3 * np.sqrt(length) # 세 배로 그리기
draw_vector(pca.mean_, pca.mean_ + v)
plt.axis('equal')
plt.show()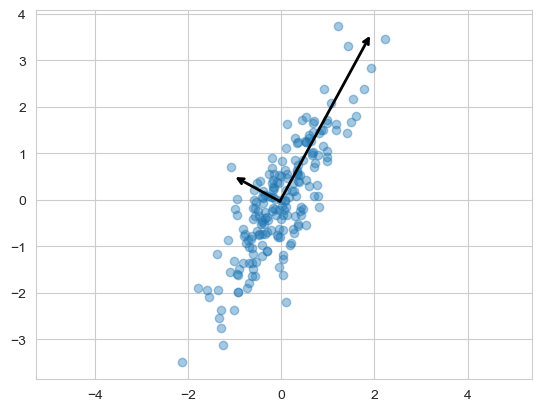
데이터의 주성분을 찾은 후 주축을 변경하는 것도 가능
pca = PCA(n_components=1, random_state=13)
pca.fit(X)n_components를 1로 두면
pca = PCA(n_components=1, random_state=13)
pca.fit(X)
X_pca = pca.transform(X)
print(pca.components_)
print(pca.explained_variance_)[[0.47802511 0.87834617]][1.82531406]
- 주요소가 하나인 데이터로 환원 (inverse transform)
- linear regression과 같은 결과일지도
X_new = pca.inverse_transform(X_pca)
plt.scatter(X[:, 0], X[:, 1], alpha=0.3)
plt.scatter(X_new[:, 0], X_new[:, 1], alpha=0.9)
plt.axis('equal')
plt.show()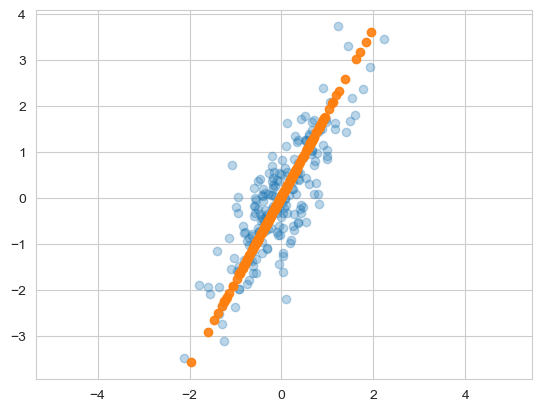
📌 실습 - iris
- PCA 에서는 scaler 적용 중요함
import pandas as pd
from sklearn.datasets import load_iris
iris = load_iris()
iris_pd = pd.DataFrame(iris.data, columns=iris.feature_names)
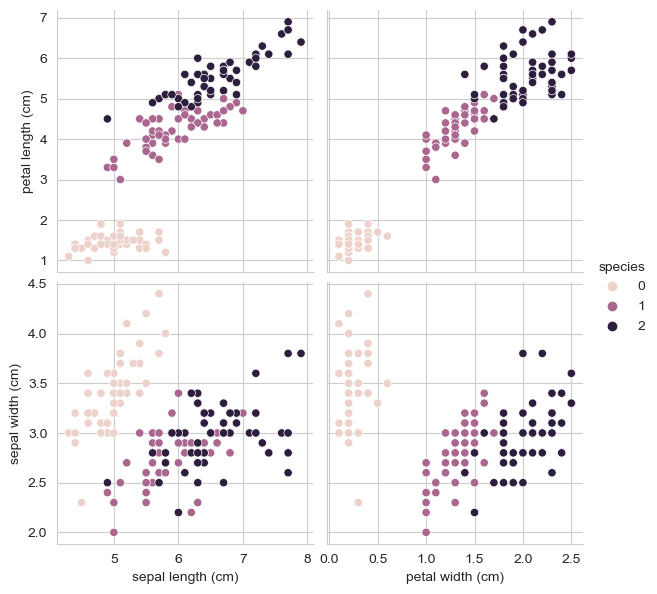
iris_pd['species'] = iris.target특성 4개를 한 번에 확인하기는 어려움
sns.pairplot(iris_pd, hue='species', height=3,
x_vars=['sepal length (cm)', 'petal width (cm)'],
y_vars=['petal length (cm)', 'sepal width (cm)']
);
from sklearn.preprocessing import StandardScaler
iris_ss = StandardScaler().fit_transform(iris.data)from sklearn.decomposition import PCA
def get_pca_data(ss_data, n_components=2):
pca = PCA(n_components=n_components)
pca.fit(ss_data)
return pca.transform(ss_data), pcairis_pca, pca = get_pca_data(iris_ss, 2)
iris_pca.shape
pca.mean_
pca.components_
pca.explained_variance_ratio_def get_pd_from_pca(pca_data, cols=['PC1', 'PC2']):
return pd.DataFrame(pca_data, columns=cols)iris_pd_pca = get_pd_from_pca(iris_pca)
iris_pd_pca['species'] = iris.target
iris_pd_pca.head()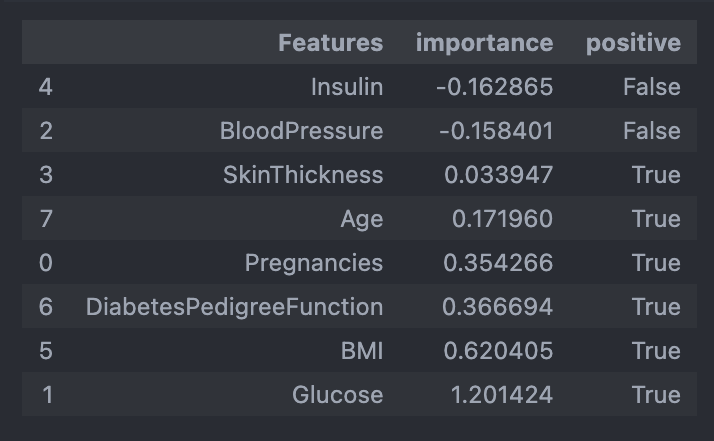
sns.pairplot(iris_pd_pca, hue='species', height=5, x_vars=['PC1'], y_vars=['PC2']);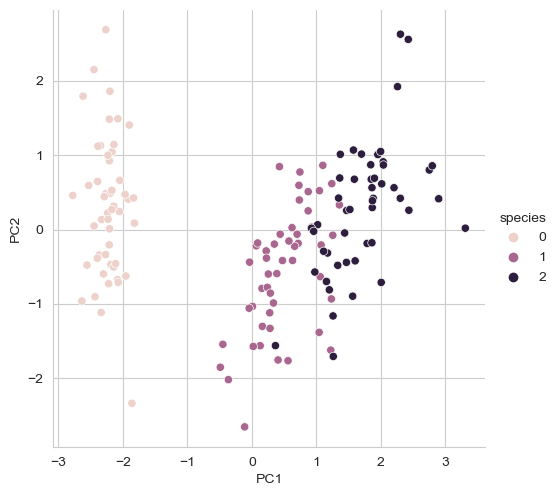
📌 Precision and Recall
와인 데이터
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=13)- 로지스틱 회귀
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
lr = LogisticRegression(solver='liblinear', random_state=13)
lr.fit(X_train, y_train)
y_pred_tr = lr.predict(X_train)
y_pred_test = lr.predict(X_test)
print('Train Acc: ', accuracy_score(y_train, y_pred_tr))
print('Test Acc: ', accuracy_score(y_test, y_pred_test))Train Acc: 0.7429286126611506
Test Acc: 0.7446153846153846
- classification report
from sklearn.metrics import classification_report
print(classification_report(y_test, lr.predict(X_test)))>>>
precision recall f1-score support
0.0 0.68 0.58 0.62 477
1.0 0.77 0.84 0.81 823
accuracy 0.74 1300
macro avg 0.73 0.71 0.72 1300
weighted avg 0.74 0.74 0.74 13000.58*(477/1300) + 0.84*(823/1300)0.7446
- confusion matrix
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, lr.predict(X_test))- precision_recall curve
import matplotlib.pyplot as plt
from sklearn.metrics import precision_recall_curve
%matplotlib inline
plt.figure(figsize=(10, 8))
pred = lr.predict_proba(X_test)[:, 1]
precisions, recalls, thresholds = precision_recall_curve(y_test, pred)
plt.plot(thresholds, precisions[:len(thresholds)], label='precision')
plt.plot(thresholds, recalls[:len(thresholds)], label='recall')
plt.grid(); plt.legend(); plt.show()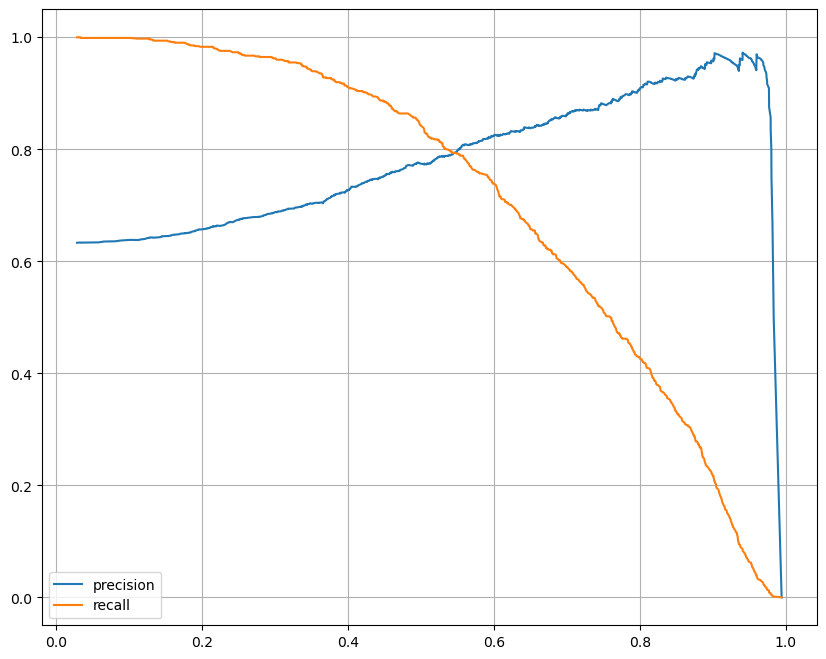
- threshold = 0.5
pred_proba = lr.predict_proba(X_test)
pred_proba[:3] # [0일 확률, 1일 확률]>>>
array([[0.40475103, 0.59524897],
[0.51010833, 0.48989167],
[0.10224708, 0.89775292]])pred_proba
>>>
array([[0.40475103, 0.59524897],
[0.51010833, 0.48989167],
[0.10224708, 0.89775292],
...,
[0.22589487, 0.77410513],
[0.67292445, 0.32707555],
[0.31386471, 0.68613529]])y_pred_test
>>>
array([1., 0., 1., ..., 1., 0., 1.])y_pred_test.reshape(-1, 1)
>>>
array([[1.],
[0.],
[1.],
...,
[1.],
[0.],
[1.]])# pred_proba 옆에 예측값 붙이기
import numpy as np
np.concatenate([pred_proba, y_pred_test.reshape(-1, 1)], axis=1)- threshold 바꿔보기 - Binarizer
from sklearn.preprocessing import Binarizer
binarizer = Binarizer(threshold=0.6).fit(pred_proba)
pred_bin = binarizer.transform(pred_proba)[:, 1]
pred_bin- classification_report
print(classification_report(y_test, lr.predict(X_test)))>>>
precision recall f1-score support
0.0 0.68 0.58 0.62 477
1.0 0.77 0.84 0.81 823
accuracy 0.74 1300
macro avg 0.73 0.71 0.72 1300
weighted avg 0.74 0.74 0.74 1300print(classification_report(y_test, pred_bin))>>>
precision recall f1-score support
0.0 0.62 0.73 0.67 477
1.0 0.82 0.74 0.78 823
accuracy 0.73 1300
macro avg 0.72 0.73 0.72 1300
weighted avg 0.75 0.73 0.74 1300