📖 앙상블 학습의 유형은 보팅(Voting), 배깅(Bagging), 부스팅(Boosting), 스태킹(Stacking) 등이 존재합니다.
이번 포스팅에서 다룰 앙상블 기법은 부스팅 알고리즘이며, 해당 알고리즘에 속하는 AdaBoost와 GBM에 대해 알아보고자 합니다.
우선 부스팅의 원리부터 차근차근 알아봅시다.
[부스팅 (Boosting)]
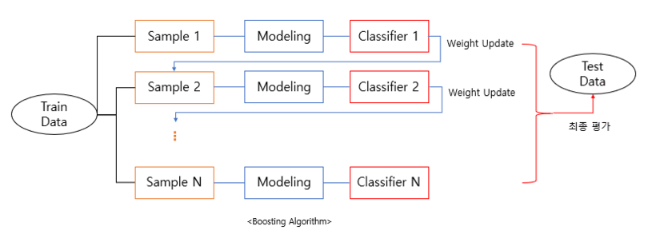
- 부스팅은 약한 학습기(weak learner)를 여러 개 연결하여 강한 학습기를 만드는 앙상블 기법입니다.
- 속도나 성능적인 측면에서 decision tree를 약한 학습기로 사용합니다.
- boosting은 bagging에서 다루었던 것처럼 복원 랜덤 샘플링을 통해 여러 모델들을 만드는 과정까지는 같으나, bagging은 병렬적으로 모델들을 결합했더라면 boosting은 단일 모델을 순차적으로 학습을 진행합니다.
- 하나의 모델을 설정하여 샘플1부터 학습을 진행합니다. 그 후, 샘플1 중에서 잘 분류하지 못한 데이터들이 있을 것입니다. 여기서 부스팅 알고리즘은 해당 데이터에 가중치를 주고 샘플2로 넘겨줍니다. 또한, 복원 랜덤 샘플링이기 때문에 샘플2에는 샘플1에 사용되지 않은 데이터도 존재할 것이고 해당 데이터에도 가중치를 적용합니다.
- 결국, 마지막으로 학습된 모델만이 아니라 이제까지 학습된 모델들을 모두 고려해서 적용하는 방식입니다.
- 이처럼 오차에 대한 가중치를 주는 방식은 정확도를 높이는 것에 큰 도움이 되지만, outlier에 취약할 수 있다는 단점이 있을 수 있습니다.
[에이다부스트 (AdaBoost)]
- Adaptive(상호보완적) Boost의 줄임말로서 일반적인 부스팅 알고리즘처럼 약한 학습기의 오류 데이터에 가중치를 부여하면서 부스팅을 수행하는 대표적인 알고리즘입니다.
- 즉, 처음 만든 데이터에서 잘못 분류한 데이터에 가중치를 증가시켜 다음 모델이 순차적으로 만들어질 때 다시 선택될 확률을 높여 더 많이 학습될 수 있도록 해줍니다. 그리고 다음 모델들이 분류를 올바르게 해내게 되면 다시 그 가중치를 감소시킵니다. 그 과정을 통해 최종 모델이 완성됩니다.
- 기존 부스팅에서는 개별 모델에서 동일한 가중치를 주었다면 adaboost는 개별 모델에 가중치를 별도로 주는 개념이 추가된 방식입니다.
- 이진 분류 문제에서 의사결정나무의 성능을 향상시키는데 가장 많이 사용됩니다.
1. 학습 방법
- 3개의 약한 학습기를 예시로 설명하겠습니다.
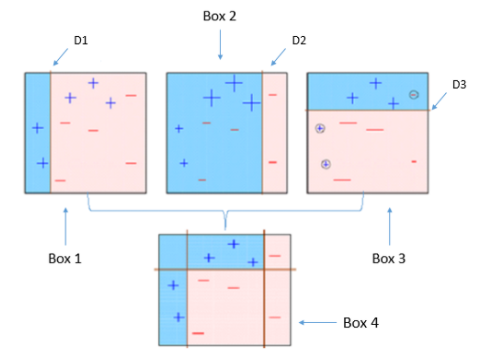
1) 첫 번째 약한 학습기가 첫 번째 분류기준(D1)으로 +와 -를 분류해서 오류 데이터를 색출
(분류는 오분류된 데이터들의 가중치 합을 최소화하는 쪽으로 진행)
2) 잘못 분류된 데이터에 대해 가중치를 부여 (커진 +표시)
3) 두 번째 약한 학습기가 가중치를 반영하여 두 번째 분류기준(D2)으로 +와 -를 다시 분류해서 오류 데이터를 색출
4) 잘못 분류된 데이터에 대해 가중치를 부여 (커진 -표시)
(오분류 데이터에는 가중치를 증가시키고 아니라면 감소시켜 데이터 가중치를 초기화)
5) 세 번째 약한 학습기가 가중치를 반영하여 세 번째 분류기준(D3)으로 +와 -를 다시 분류해서 오류 데이터를 색출
6) 마지막으로 3개의 분류기준별로 계산된 모델의 가중치를 각각 부여하고 가중합으로 결합하여 분류
2. 장점
- 과적합되는 경향을 줄일 수 있습니다.
- Random Forest와 비교했을 때 이진분류에서 대체적으로 결과가 더 좋게 나옵니다.
3. 단점
- 노이즈 데이터 및 이상치에 민감합니다.
4. 실습
> library(caret)
> # ['ada'가 포함된 모델 확인]
> grep('ada', names(getModelInfo()), value = T, ignore.case = T)
[1] "ada" "AdaBag" "AdaBoost.M1" "adaboost" "mxnetAdam"
> # [adaboost의 파라미터 확인]
> modelLookup('adaboost')
model parameter label forReg forClass probModel
1 adaboost nIter #Trees FALSE TRUE TRUE
2 adaboost method Method FALSE TRUE TRUE
> # [최적의 모수를 찾기 위한 학습방법 사전 설정]
> control <- caret::trainControl(method = 'repeatedcv',
+ search = 'random',
+ ## 하이퍼파라미터 random search
+ number = 3,
+ repeats = 3,
+ allowParallel = T,
+ verboseIter = T
+ )
> # [모델 학습 및 검증]
> ada_model <- train(credit.rating ~., train,
+ method = "adaboost",
+ metric = 'Accuracy',
+ preProcess = c("zv", "center", "scale", "spatialSign"),
+ # tuneLength = 7,
+ trControl = control)
+ Fold1.Rep1: nIter=116, method=Adaboost.M1
- Fold1.Rep1: nIter=116, method=Adaboost.M1
+ Fold1.Rep1: nIter=506, method=Adaboost.M1 ...
> # [최적모델 도출]
> ada_model
AdaBoost Classification Trees
700 samples
20 predictor
2 classes: 'pos', 'neg'
Pre-processing: centered (20), scaled (20), spatial sign
transformation (20)
Resampling: Cross-Validated (3 fold, repeated 3 times)
Summary of sample sizes: 466, 467, 467, 467, 467, 466, ...
Resampling results across tuning parameters:
nIter method Accuracy Kappa
25 Real adaboost 0.7242785 0.2222041
207 Adaboost.M1 0.7385724 0.3222722
672 Adaboost.M1 0.7419085 0.3356293
Accuracy was used to select the optimal model using the largest value.
The final values used for the model were nIter = 672 and method
= Adaboost.M1.
> # [변수 중요도 확인]
> plot(varImp(ada_model))
> # [혼돈행렬]
> confusionMatrix(test$credit.rating, predict(ada_model, test, type = 'raw'))
Confusion Matrix and Statistics
Reference
Prediction pos neg
pos 39 51
neg 24 186
Accuracy : 0.75
95% CI : (0.697, 0.798)
No Information Rate : 0.79
P-Value [Acc > NIR] : 0.95956
Kappa : 0.349
Mcnemar's Test P-Value : 0.00268
Sensitivity : 0.6190
Specificity : 0.7848
Pos Pred Value : 0.4333
Neg Pred Value : 0.8857
Prevalence : 0.2100
Detection Rate : 0.1300
Detection Prevalence : 0.3000
Balanced Accuracy : 0.7019
'Positive' Class : pos [그래디언트 부스팅 머신 (GBM)]
-
앞선 adaBoost의 경우 오분류한 데이터에 가중치를 줌으로써 모델을 보완했지만, GBM의 경우 가중치 업데이트를 경사하강법(Gradient Descent)을 이용하여 최적화된 결과를 얻는 알고리즘입니다.
- 경사하강법은 loss fuction을 정의하고 이의 미분값이 최소가 되도록 하는 방향을 찾고 접근하는 방식입니다. 예를 들어 loss function을 squared error로 정의한다면 아래와 같은 식으로 표현할 수 있습니다. 이를 미분한다면 기울기 값을 구할 수 있고 이것을 최소로 만드는 것이 목적입니다.
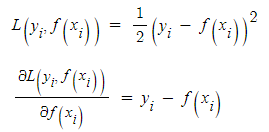
- 편미분을 통해 얻은 gradient는 결국 f(x)가 loss를 줄이기 위해 가야하는 방향인데 식을 보시다시피 residual이라는 것을 알 수 있습니다.
- 경사하강법은 loss fuction을 정의하고 이의 미분값이 최소가 되도록 하는 방향을 찾고 접근하는 방식입니다. 예를 들어 loss function을 squared error로 정의한다면 아래와 같은 식으로 표현할 수 있습니다. 이를 미분한다면 기울기 값을 구할 수 있고 이것을 최소로 만드는 것이 목적입니다.
-
AdaBoost처럼 반복마다 샘플의 가중치를 조정하는 대신 이전 예측기가 만든 잔여 오차(residual error)에 새로운 예측기를 학습시킵니다.
-
이전 모델의 residual을 가지고 weak learner를 강화합니다. 즉, residual을 예측하는 형태의 모델입니다.
1. 학습 방법
- 수치형 반응변수를 예측하는 회귀 문제에서, 3개의 모델을 이용하여 예시를 들어보겠습니다.
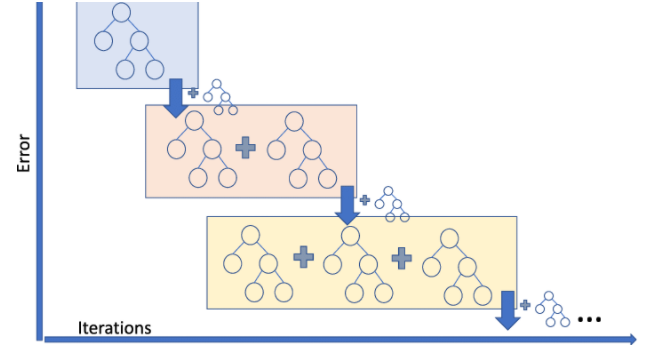
1) 먼저 A라는 모델은 우선 전체 데이터의 target variable의 평균으로 예측값을 만듭니다.
이 후, 실제값 - (A모델의 예측값(=평균값) = 잔차(residual)를 구합니다.
2) 이제 B모델은 A모델의 학습에 사용했던 features를 가지고 residual을 맞추는 방식으로 학습을 진행합니다.
그리고 실제값 - (A모델의 예측값(=평균값) + learning_rateB모델의 예측값) = 새로운 residual을 구합니다.
3) 그 다음 C모델도 마찬가지로, A모델의 학습에 사용했던 features를 가지고 바로 위에서 언급한 새로운 residual을 맞추도록 학습합니다.
그리고 실제값 - (A모델의 예측값 + learning_rateB모델의 예측값 + learning_rate*C모델의 예측값) = 새로운 residual을 구합니다.
4) 그 다음 모델이 있다면 다시 새로운 residual을 학습하게 됩니다.
구체적인 학습 프로세스 참고
2. 장점
- ML 계열의 모델 중 성능이 좋은 편에 속합니다.
3. 단점
- 수행시간이 오래걸립니다.
- 하이퍼파라미터 튜닝 노력이 필요합니다.
- 탐욕적 알고리즘(Greedy Algorithm)으로 과적합이 빠르게 진행될 수 있습니다.
- 탐욕적 알고리즘이란, 미래를 생각하지 않고 각 단계에서 가장 최선의 선택을 하는 기법을 말합니다.
4. 실습
> # [최적의 모수를 찾기 위한 학습방법 사전 설정]
> fitControl <- trainControl(method = "repeatedcv",
+ number = 3,
+ repeats = 3,
+ verboseIter = T)
> # [gbm의 파라미터 확인]
> modelLookup('gbm')
model parameter label forReg forClass
1 gbm n.trees # Boosting Iterations TRUE TRUE
2 gbm interaction.depth Max Tree Depth TRUE TRUE
3 gbm shrinkage Shrinkage TRUE TRUE
4 gbm n.minobsinnode Min. Terminal Node Size TRUE TRUE
probModel
1 TRUE
2 TRUE
3 TRUE
4 TRUE
> # [그리드서치를 위한 하이퍼파라미터 설정]
> tunegrid2 <- expand.grid(n.trees = c(10, 20, 30, 40),
+ ## 생성할 나무의 개수
+ interaction.depth = c(1:10),
+ ## = maxdepth
+ shrinkage = c(0.1),
+ #=learning Rate
+ n.minobsinnode = c(10:50)
+ ## 분할을 시작할 노드의 최소 훈련 세트 샘플 수
+ )
> # [모델 학습 및 검증]
> gbm_gridsearch2 <- train(credit.rating~.,
+ data = train,
+ method = 'gbm',
+ metric = ifelse(is.factor(train$credit.rating),'Accuracy','RMSE'),
+ tuneGrid = tunegrid2,
+ trControl = fitControl)
> # [파라미터에 따른 accuracy 흐름 파악]
> trellis.par.set(caretTheme())
> plot(gbm_gridsearch2)
> # [변수 중요도 확인]
> summary(gbm_gridsearch2)
var rel.inf
account.balance account.balance 21.1839170
credit.amount credit.amount 19.3042177
credit.duration.months credit.duration.months 10.8977305
credit.purpose credit.purpose 6.9162193
age age 6.9126727
savings savings 5.8127936
previous.credit.payment.status previous.credit.payment.status 5.6003470
current.assets current.assets 5.5444213
installment.rate installment.rate 4.1435804
employment.duration employment.duration 3.6837592
marital.status marital.status 3.1228349
residence.duration residence.duration 1.9844654
apartment.type apartment.type 1.9317795
other.credits other.credits 1.2082644
dependents dependents 1.0179670
telephone telephone 0.4575234
occupation occupation 0.2775065
guarantor guarantor 0.0000000
bank.credits bank.credits 0.0000000
foreign.worker foreign.worker 0.0000000
> # [혼돈행렬]
> confusionMatrix(test$credit.rating, predict(gbm_gridsearch2, test))
Confusion Matrix and Statistics
Reference
Prediction pos neg
pos 40 50
neg 23 187
Accuracy : 0.7567
95% CI : (0.704, 0.8041)
No Information Rate : 0.79
P-Value [Acc > NIR] : 0.929670
Kappa : 0.3663
Mcnemar's Test P-Value : 0.002342
Sensitivity : 0.6349
Specificity : 0.7890
Pos Pred Value : 0.4444
Neg Pred Value : 0.8905
Prevalence : 0.2100
Detection Rate : 0.1333
Detection Prevalence : 0.3000
Balanced Accuracy : 0.7120
'Positive' Class : pos 