📖 이번 포스팅은 이전 포스트처럼 고려대학교 김성범 교수님의 강의영상을 참고하여 모델 기반 이상치 탐지 알고리즘 중 Isolation Forest에 대해 다루고자 합니다.
Isolation Forest
Notion
- 정상 데이터로부터 학습한 모델을 기반으로 각 객체의 정상/이상 여부를 판단하는 방법론입니다.
- 여러 개의 의사결정나무를 종합한 앙상블 기반의 이상탐지 기법으로 의사결정나무를 지속적으로 분기시키면서 모든 데이터 관측치의 고립 정도 여부에 따라 이상치를 판별합니다.
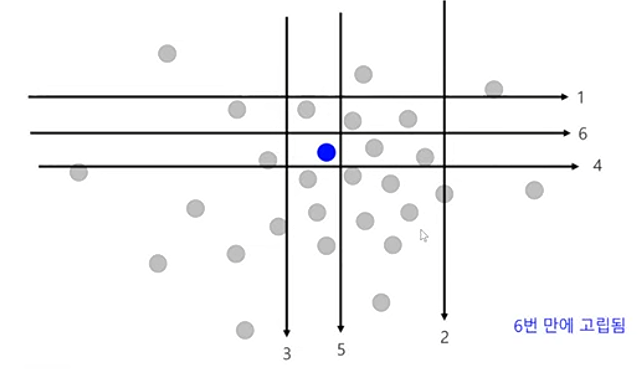
- 정상 관측치는 고립되기 어려울 것이다라는 가정을 합니다. 이는 정상 관측치를 고립시키기 위해서는 2진 분할을 여러 번 수행해야 한다는 것입니다.
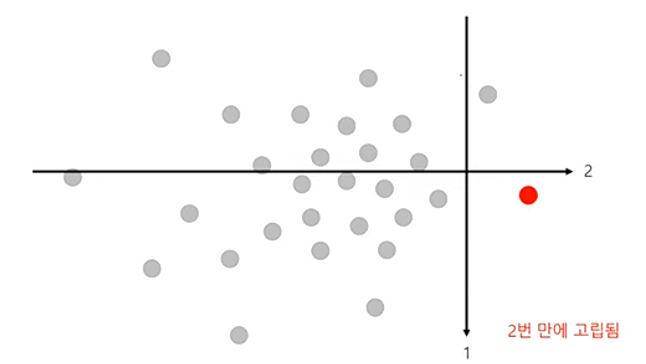
- 이상치는 쉽게 고립될 것이다라는 가정을 합니다. 이는, 이상치를 고립시키기 위해서는 2진 분할을 적게 수행해야 한다는 것입니다.
- 직관적으로 비정상 데이터라면 의사결정나무의 루트 노드에서 가까운 깊이에서 고립될 것이고, 정상 데이터라면 루트 노드에서 먼 깊이에서 고립될 것입니다.
- 각 관측치의 Path length(관측치 x가 고립될 때까지 필요한 분할 횟수)를 기반으로 Anomaly score를 정의하여 부여합니다.
- 2008년에 발표된 모델이지만 아직까지도 이상치 탐지를 하는데 있어 유용하게 사용됩니다.
Algorithm
1. 전체 데이터에서 일부 관측치를 랜덤하게 선택
2. 랜덤하게 선택된 관측치에 대해 임의의 변수(splitting variable)와 분할점(splitting point)을 사용하여 다음 조건을 만족할 때까지 이진분할 진행
- 의사결정나무모델이 사전 정의된 깊이에 도달
- 모든 터미널 노드에 존재하는 관측치가 1개씩 존재
- 모든 터미널 노드에 존재하는 관측치들이 같은 입력변수
3. 위와 같은 과정으로 여러 개의 ITree를 구축
4. ITree마다 각 관측치의 Path length(분리 횟수)를 저장
5. 각 관측치의 평균 Path length를 기반으로 Anomaly score(이상치 스코어)를 계산 및 이상치 판별
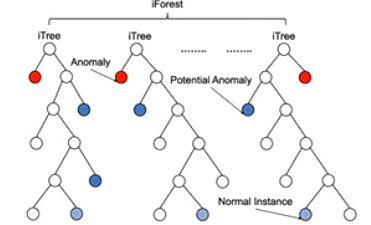
Anomaly score 정의
-
Path length를 기반으로 정의
-
Path length
-
Normalized (모든 관측치들의 평균 Path length)
-
Anomaly score :
- = 모든 관측치들의 평균 Path length 대비 내가 관심있는 관측치 평균 Path length 도출
-
(Path length의 평균값이 0에 가까움으로 이상 데이터) -
(Path length의 평균값이 n에 가까움으로 정상 데이터) -
(Path length의 평균값이 normalized 와 비슷함으로 정상 데이터)
- Anomaly score의 범위는 0~1이며, 1에 가까우면 이상 데이터, 0.5이하면 정상 데이터라고 할 수 있습니다.
Code

- 파이썬에서 sklearn.ensemble 모듈의 IsolationForest함수가 해당 알고리즘을 지원합니다.
- 일반적인 sklearn의 다른 함수와 마찬가지로 데이터에 'fit()', 'predict()'를 통해 결과를 알 수 있으며, 'predict()'의 결과는 정상(1), 이상(-1)로 표현됩니다.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set_style("darkgrid")
from sklearn.ensemble import IsolationForest
rng = np.random.RandomState(42)
# Generating training data
X_train = 0.2 * rng.randn(1000, 2)
X_train = np.r_[X_train + 3, X_train]
X_train = pd.DataFrame(X_train, columns = ['x1', 'x2'])
# Generating new, 'normal' observation
X_test = 0.2 * rng.randn(200, 2)
X_test = np.r_[X_test + 3, X_test]
X_test = pd.DataFrame(X_test, columns = ['x1', 'x2'])
# Generating outliers
X_outliers = rng.uniform(low=-1, high=5, size=(50, 2))
X_outliers = pd.DataFrame(X_outliers, columns = ['x1', 'x2'])
# data scatter
plt.rcParams['figure.figsize'] = [10, 10]
p1 = plt.scatter(X_train.x1, X_train.x2, c='white', s=20*4, edgecolor='k', label='training observations')
# p2 = plt.scatter(X_test.x1, X_test.x2, c='green', s=20*4, edgecolor='k', label='new regular obs.')
p3 = plt.scatter(X_outliers.x1, X_outliers.x2, c='red', s=20*4, edgecolor='k', label='new abnormal obs.')
plt.legend()
# predict
clf = IsolationForest(max_samples=100, contamination = 0.1, random_state=42)
## 전체 데이터 비율의 10%를 이상치로 지정
clf.fit(X_train)
y_pred_train = clf.predict(X_train)
y_pred_test = clf.predict(X_test)
y_pred_outliers = clf.predict(X_outliers)
# outliers예측값과 X_outliers값을 X_outliers2변수에 할당
X_outliers2 = X_outliers.assign(y = y_pred_outliers)
X_outliers2
# 이상 탐지 scatter
plt.figure(figsize = (12,10))
p1 = plt.scatter(X_train.x1, X_train.x2, c='white',
s=20*4, edgecolor='k', label="training observations")
p2 = plt.scatter(X_outliers2.loc[X_outliers2.y == -1, ['x1']],
X_outliers2.loc[X_outliers2.y == -1, ['x2']],
c='red', s=20*4, edgecolor='k', label="detected outliers")
p3 = plt.scatter(X_outliers2.loc[X_outliers2.y == 1, ['x1']],
X_outliers2.loc[X_outliers2.y == 1, ['x2']],
c='green', s=20*4, edgecolor='k', label="detected regular obs")
plt.legend()
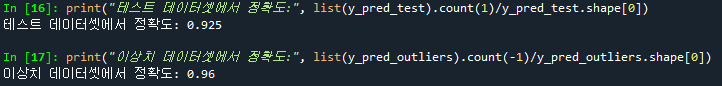
print("테스트 데이터셋(normal data)에서 정확도:", list(y_pred_test).count(1)/y_pred_test.shape[0])
print("이상치 데이터셋에서 정확도:", list(y_pred_outliers).count(-1)/y_pred_outliers.shape[0])
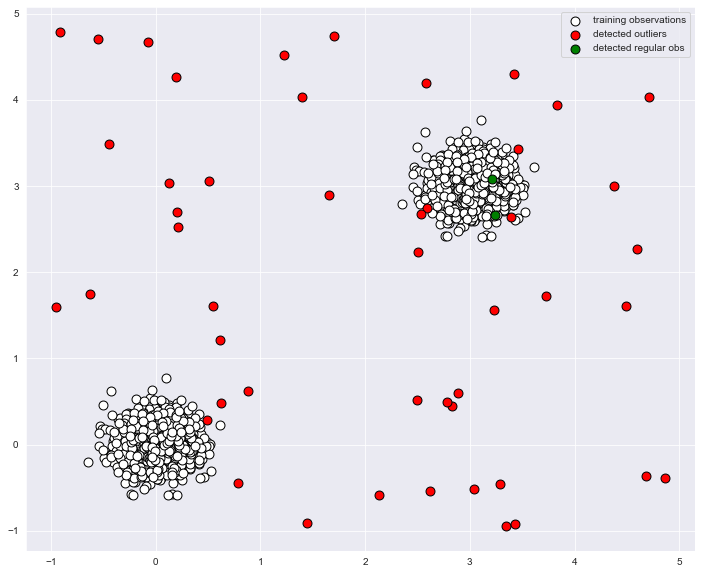

기존 데이터에서 그 데이터가 진짜 이상치인지를 알 수가 없는데 어떻게 정확도를 구할 수 있는 건가요? list(y_pred_test).count(1)/y_pred_test.shape[0]) 이게 왜 정확도인건지 궁금합니다.