Random Forest Classifier
Introduction
The Random Forest Classifier is a powerful and versatile machine learning algorithm used for classification tasks. It operates by constructing a multitude of decision trees at training time and outputting the class that is the mode of the classes (classification) of the individual trees. Random forests correct for decision trees' habit of overfitting to their training set, providing a more generalized model.
Background and Theory
The algorithm was first introduced by Leo Breiman and Adele Cutler in 2001. Random Forests belong to the ensemble learning family, which means they use multiple learning algorithms to obtain better predictive performance than could be obtained from any of the constituent learning algorithms alone.
A Random Forest Classifier builds multiple decision trees and merges them together to get a more accurate and stable prediction. The core concept behind random forests is the idea of "bagging" or Bootstrap Aggregating, where multiple models (in this case, trees) are trained on different parts of the same training set and then averaged to improve the stability and accuracy.
Mathematical Foundations
Each tree in the forest is built from a sample drawn with replacement (i.e., a bootstrap sample) from the training set. Additionally, when splitting a node during the construction of the tree, the split that is chosen is no longer the best split among all features. Instead, the split that is picked is the best split among a random subset of the features. As a result, the correlation between trees in the forest is decreased, leading to a decrease in the forest's variance without an increase in bias.
Procedural Steps
- Bootstrap sampling: Randomly select N samples from the training data with replacement to create a subset of the data. This is done for each tree to be trained.
- Tree building: For each bootstrap sample, grow a decision tree. At each node:
- Randomly select features from the total features.
- Choose the best split from these features to split the node.
- Split the node into child nodes.
- Repeat the process for a specified number of trees.
- Voting for classification: For classification tasks, each tree votes, and the most popular class is chosen as the final outcome for the input data.
Mathematical Formulations
Given a training set with corresponding labels , a random forest classifier constructs a collection of decision trees , each trained on a bootstrap sample of the training data.
The prediction of the random forest, , for an input sample is given by:
where denotes the statistical mode (i.e., the most frequent label among the predictions of the individual trees).
Implementation
Parameters
n_trees:int, default = 100
Number of trees in the forest
max_depth:int, default = 10
Maximum depth of each trees
criterion:Literal['gini', 'entropy'], default = ‘gini’
Function used to measure the quality of a split
min_samples_split:int, default = 2
Minimum samples required to split a node
min_samples_leaf:int, default = 1
Minimum samples required to be at a leaf node
max_features:int, default = None
Number of features to consider
min_impurity_decrease:float, default = 0.0
Minimum decrement of impurity for a split
max_leaf_nodes:int, default = 1
Maximum amount of leaf nodes
bootstrap:bool, default = True
Whether to bootstrap the samples of dataset
bootstrap_feature:bool, default = False
Whether to bootstrap the features of each data
n_features:intLiteral['auto'], default = ‘auto’
Number of features to be sampled when bootstrapping features
Examples
Test on standardized and reduced() digits dataset with only 5 classes:
from luma.ensemble.forest import RandomForestClassifier
from luma.preprocessing.scaler import StandardScaler
from luma.model_selection.split import TrainTestSplit
from luma.reduction.linear import KernelPCA
from luma.visual.evaluation import DecisionRegion, ConfusionMatrix
from sklearn.datasets import load_digits
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(42)
X, y = load_digits(n_class=5, return_X_y=True)
indices = np.random.choice(X.shape[0], size=500)
X_sample = X[indices]
y_sample = y[indices]
sc = StandardScaler()
X_sample_std = sc.fit_transform(X_sample)
kpca = KernelPCA(n_components=2, gamma=0.01, kernel='rbf')
X_kpca = kpca.fit_transform(X_sample_std)
X_train, X_test, y_train, y_test = TrainTestSplit(X_kpca, y_sample,
test_size=0.3).get
forest = RandomForestClassifier(n_trees=10,
max_depth=100,
criterion='gini',
min_impurity_decrease=0.01,
bootstrap=True)
forest.fit(X_train, y_train)
X_cat = np.concatenate((X_train, X_test))
y_cat = np.concatenate((y_train, y_test))
score = forest.score(X_cat, y_cat)
fig = plt.figure(figsize=(10, 5))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
dec = DecisionRegion(forest, X_cat, y_cat, cmap='Spectral')
dec.plot(ax=ax1)
ax1.set_title(f'RandomForestClassifier [Acc: {score:.4f}]')
conf = ConfusionMatrix(y_cat, forest.predict(X_cat), cmap='BuPu')
conf.plot(ax=ax2, show=True)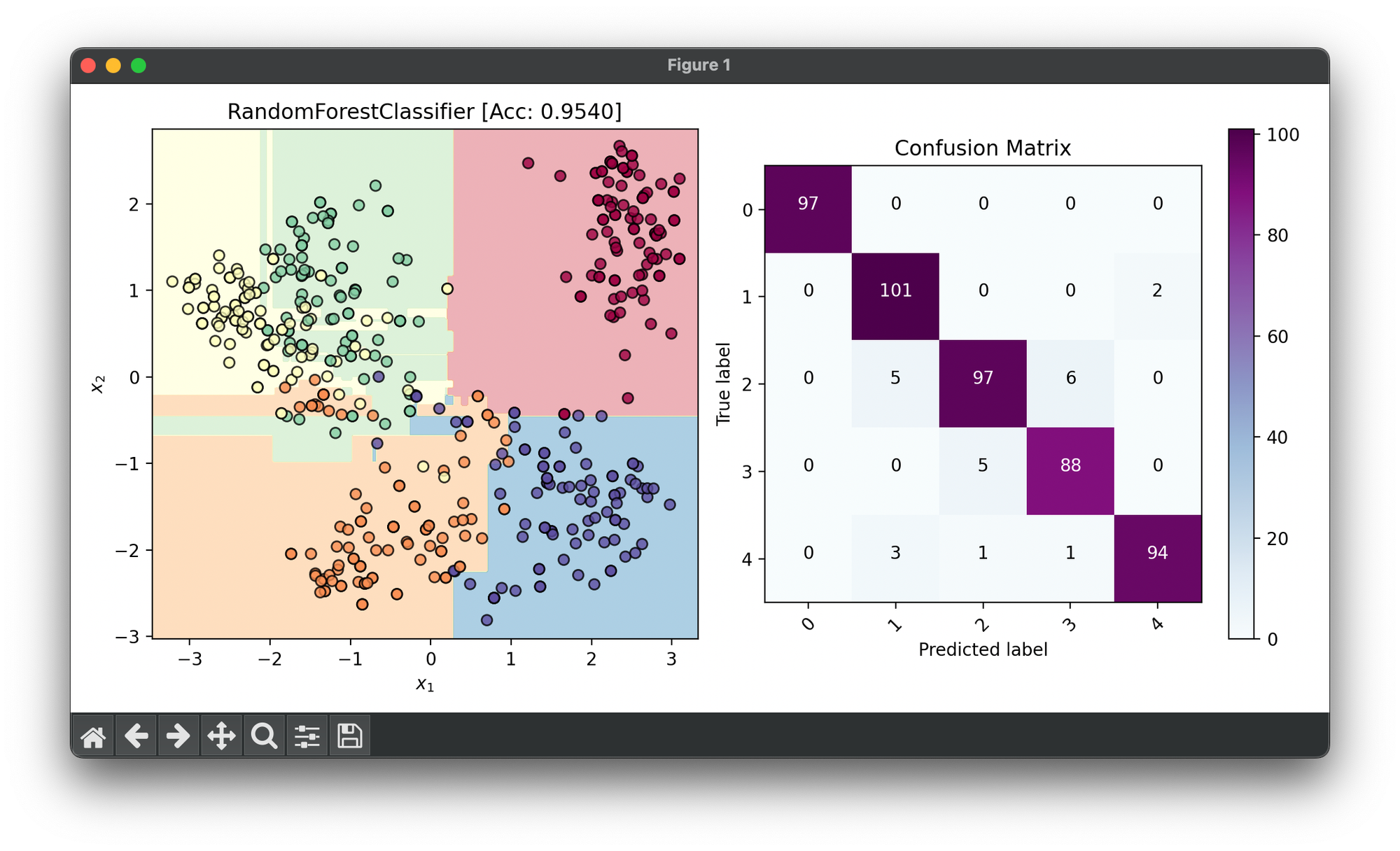
Applications
- Medical Diagnosis: Random Forests are used for diagnosing diseases by analyzing patient data.
- Financial Market Analysis: They can predict stock market movements by analyzing historical data.
- E-Commerce: For recommendation systems, determining a customer's likelihood to purchase a product.
- Fraud Detection: Identifying fraudulent activities in banking and insurance.
Strengths and Limitations
Strengths
- Versatility: Can be used for both classification and regression tasks.
- Handles Large Data Sets: Efficiently processes large databases.
- Automatic Feature Selection: Offers insights into feature importance.
Limitations
- Complexity: More complex and computationally intensive than decision trees.
- Model Interpretability: The model is not as easy to interpret as a single decision tree.
- Overfitting with Noisy Data: While it is resistant to overfitting, random forests can still overfit noisy datasets.
Advanced Topics
- Feature Importance: Random Forests can be used to rank the importance of variables in a regression or classification problem.
- Handling Missing Values: They can handle missing data by surrogate splits in the trees.
- Parallelization: Training of the trees can be easily parallelized, as each tree is built independently.
References
- Breiman, L. (2001). Random forests. Machine learning, 45(1), 5-32.
- Liaw, A., & Wiener, M. (2002). Classification and regression by randomForest. R News, 2(3), 18-22.
