Chapter 15 주성분 분석
-
PCA: 정보를 최대한 보존하면서 새로운 축을 찾아, 고차원을 저차원으로 변환
-
차원축소와 변수추출 기법으로 많이 쓰이는 주성분 분석
-
데이터를 어떤 벡터에 정사영시켜 차원을 낮출 수 있음

-
새로운 특성을 찾아서 표현하는 것
from sklearn.decomposition import PCA
pca = PCA(n_components=2, random_state=13) # 주성분을 2개만 찾아라
pca.fit(X) def draw_vector(v0, v1, ax=None):
ax = ax or plt.gca()
arrowprops = dict(arrowstyle='->', linewidth=2, color='black', shrinkA=0, shrinkB=0)
ax.annotate('', v1, v0, arrowprops=arrowprops) plt.scatter(X[:, 0], X[:, 1], alpha=0.4)
for length, vector in zip(pca.explained_variance_, pca.components_):
v = vector * 3 * np.sqrt(length)
draw_vector(pca.mean_, pca.mean_ + v)
plt.axis('equal');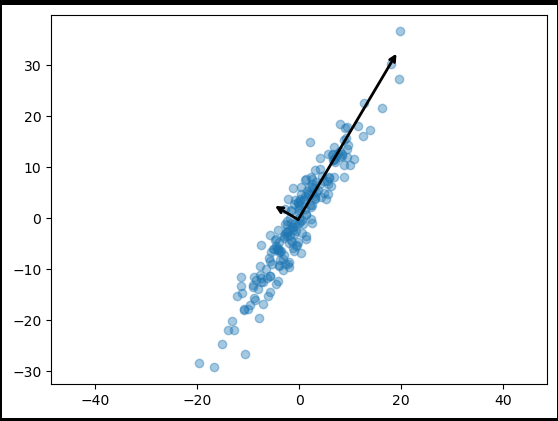
pca = PCA(n_components=1, random_state=13)
pca.fit(X)
X_pca = pca.transform(X)
pca.explained_variance_ratio_X_new = pca.inverse_transform(X_pca)
plt.scatter(X[:, 0], X[:, 1], alpha=0.4)
plt.scatter(X_new[:, 0], X_new[:, 1], alpha=0.8)
plt.axis('equal');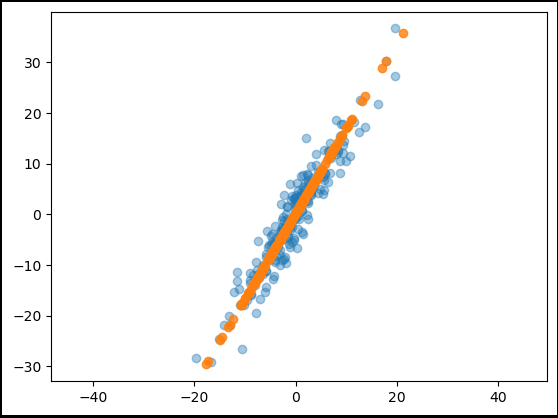
Iris 데이터에 PCA 적용
import pandas as pd
from sklearn.datasets import load_iris
iris = load_iris()
iris_pd = pd.DataFrame(iris.data, columns=iris.feature_names)
iris_pd['species'] = iris.target
iris_pd.head()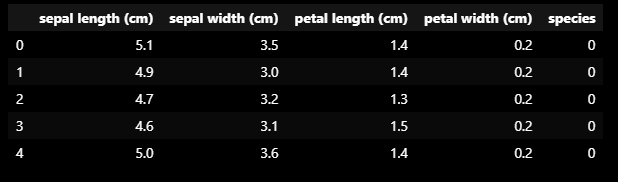
- Iris 데이터는 특성이 4개라 한번에 그릴 수 없음
from sklearn.preprocessing import StandardScaler
iris_ss = StandardScaler().fit_transform(iris.data)
iris_ss[:5]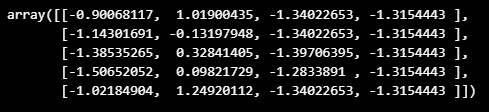
from sklearn.decomposition import PCA
def get_pca_data(ss_data, n_components=2):
pca = PCA(n_components=n_components)
pca.fit(ss_data)
return pca.transform(ss_data), pca iris_pca, pca = get_pca_data(iris_ss, 2)
iris_pca.shape(150, 2)
pca.mean_array([-1.69031455e-15, -1.84297022e-15, -1.69864123e-15, -1.40924309e-15])
pca.components_ array([[ 0.52106591, -0.26934744, 0.5804131 , 0.56485654],
[ 0.37741762, 0.92329566, 0.02449161, 0.06694199]])
def get_pd_from_pca(pca_data, cols=['PC1', 'PC2']):
return pd.DataFrame(pca_data, columns=cols)
iris_pd_pca = get_pd_from_pca(iris_pca)
iris_pd_pca['species'] = iris.target
iris_pd_pca.head() 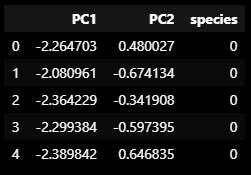
sns.pairplot(iris_pd_pca, hue='species'); 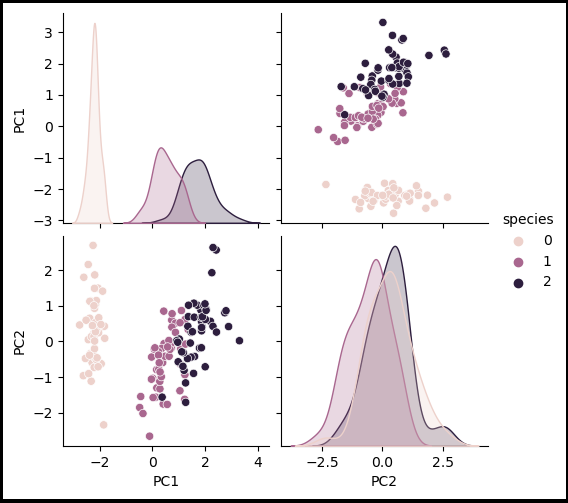
def print_variance_ratio(pca):
print('Explained variance ratio: {}'.format(pca.explained_variance_ratio_))
print('Cumulative explained variance ratio: {}'.format(np.cumsum(pca.explained_variance_ratio_)))
print_variance_ratio(pca) Explained variance ratio: [0.72962445 0.22850762]
Cumulative explained variance ratio: [0.72962445 0.95813207]
와인 데이터에 PCA 적용
wine_ss = StandardScaler().fit_transform(wine_X)
pca_wine, pca = get_pca_data(wine_ss, 2)
pca_wine.shape (6497, 2)
print_variance_ratio(pca) Explained variance ratio: [0.25346226 0.22082117]
Cumulative explained variance ratio: [0.25346226 0.47428343]
pca_columns = ['PC1', 'PC2']
pca_wine_pd = get_pd_from_pca(pca_wine, pca_columns)
pca_wine_pd['color'] = wine_y.values
sns.pairplot(pca_wine_pd, hue='color'); 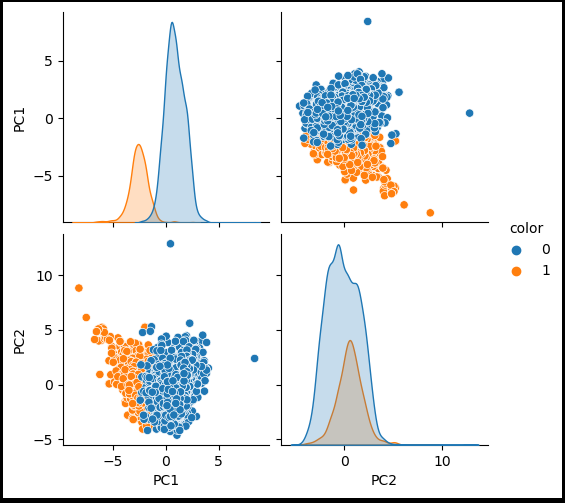
pca_wine, pca = get_pca_data(wine_ss, 3)
print_variance_ratio(pca) Explained variance ratio: [0.25346226 0.22082117 0.13679223]
Cumulative explained variance ratio: [0.25346226 0.47428343 0.61107566]
import plotly.express as px
fig = px.scatter_3d(pca_wine_plot, x='PC1', y='PC2', z='PC3', color='color')
fig.update_layout(margin=dict(l=0, r=0, b=0, t=0))
fig.show() 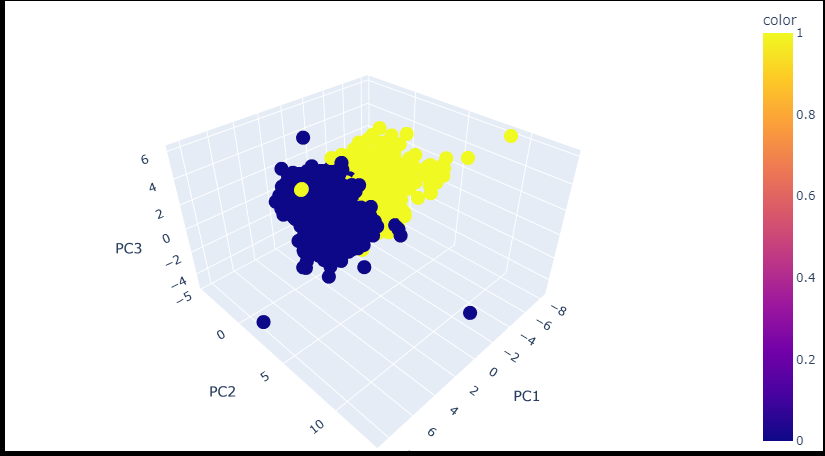
- 주성분 분석은 데이터를 시각화 하는 부분에도 도움을 줌
이 글은 제로베이스 데이터 취업 스쿨의 강의 자료 일부를 발췌하여 작성되었습니다