Locally Linear Embedding (LLE)
Introduction
Locally Linear Embedding (LLE) is a non-linear dimensionality reduction technique widely used for exploring the structure of high-dimensional data. The key idea behind LLE is to preserve local neighborhoods within the data, making it particularly effective for uncovering the underlying manifold structure. This technique is well-suited for tasks where the data lie on a well-defined manifold in a high-dimensional space, and the goal is to project this data onto a lower-dimensional space for visualization, analysis, or further processing.
Background and Theory
LLE starts with the assumption that each high-dimensional data point and its nearest neighbors lie on or close to a locally linear patch of the manifold. The algorithm consists of two main steps: First, it computes linear coefficients that best reconstruct each data point from its neighbors, preserving the local geometric configurations. Second, it uses these coefficients to compute a low-dimensional embedding of the data that best preserves these local properties.
Mathematical Foundations
Given a set of data points where each (with being the dimensionality of the input space), LLE seeks to find a set of points in a lower-dimensional space (, ) such that each best represents based on its neighbors.
Reconstruction Weights
The first step involves computing the weights that best linearly reconstruct each point from its nearest neighbors. This is done by minimizing the reconstruction error:
subject to the constraint for all , where if is not a neighbor of .
Embedding
The second step aims to find the low-dimensional embeddings that preserve the local geometry represented by the weights . This involves minimizing the embedding cost function:
with respect to , given the weights computed in the first step.
Procedural Steps
- Neighbor Selection: For each data point , identify its nearest neighbors.
- Weight Matrix Construction: Solve for the weights that best reconstruct each point from its neighbors, with the constraint that the weights for each point sum to one.
- Low-Dimensional Embedding: Using the weights , compute the low-dimensional representations that best preserve the local configurations.
Implementation
Parameters
n_components:int, default = None
Dimensionality of low-space
n_neighbors:int, default = 5
Number of neighbors to be considered 'close’
Examples
Test on the 3D Swiss-roll dataset:
from luma.reduction.manifold import LLE
from sklearn.datasets import make_swiss_roll
import matplotlib.pyplot as plt
X, y = make_swiss_roll(n_samples=500, noise=0.5)
model = LLE(n_components=2, n_neighbors=7)
Z = model.fit_transform(X)
fig = plt.figure(figsize=(10, 5))
ax1 = fig.add_subplot(1, 2, 1, projection='3d')
ax2 = fig.add_subplot(1, 2, 2)
ax1.scatter(X[:, 0], X[:, 1], X[:, 2], c=y, cmap='rainbow')
ax1.set_xlabel(r'$x$')
ax1.set_ylabel(r'$y$')
ax1.set_zlabel(r'$z$')
ax1.set_title('Original Swiss Roll')
ax2.scatter(Z[:, 0], Z[:, 1], c=y, cmap='rainbow')
ax2.set_xlabel(r'$z_1$')
ax2.set_ylabel(r'$z_2$')
ax2.set_title(f'After {type(model).__name__}')
ax2.grid(alpha=0.2)
plt.tight_layout()
plt.show()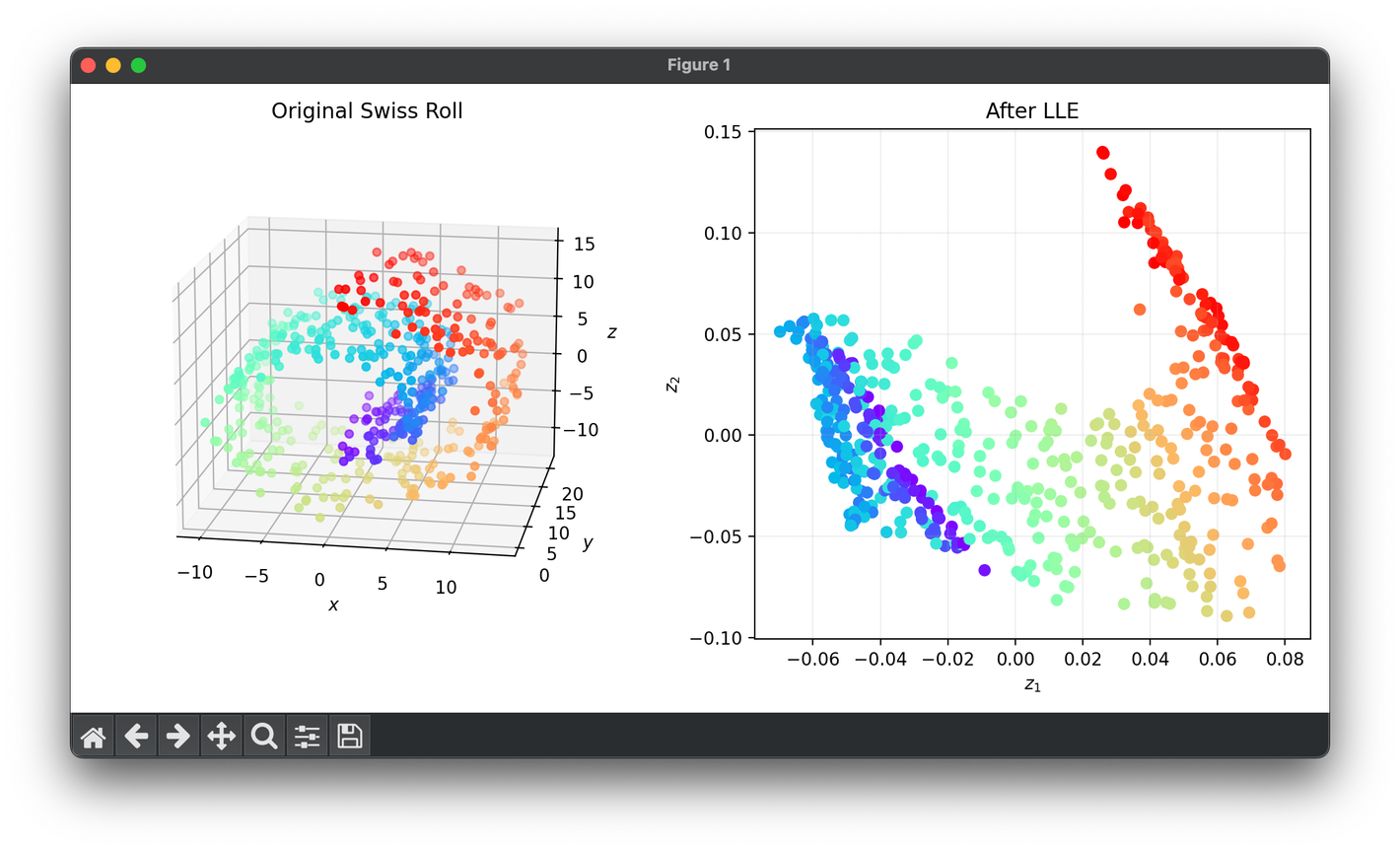
Applications
LLE is applied in a variety of fields, including computer vision, signal processing, bioinformatics, and machine learning, particularly for tasks like data visualization, noise reduction, and feature extraction.
Strengths and Limitations
Strengths
- Non-linearity: Effective at uncovering the non-linear structure of data.
- Manifold Learning: Excels at learning the manifold structure, especially in cases where the manifold is well-defined and smooth.
Limitations
- Sensitivity to Parameters: The choice of the number of neighbors () can significantly affect the outcome.
- Data Density: Performs poorly on data with varying density.
- Optimization Challenges: Finding the global minimum of the embedding cost function can be challenging.
Advanced Topics
- Improvements and Variants: Several variants and improvements of LLE have been proposed, including modified objective functions and algorithms that better handle data with varying density or noise.
- Combination with Other Techniques: LLE can be combined with other dimensionality reduction or machine learning techniques for enhanced performance in specific tasks.
References
- Roweis, S. T., & Saul, L. K. "Nonlinear Dimensionality Reduction by Locally Linear Embedding." Science, vol. 290, no. 5500, 2000, pp. 2323-2326.
- Saul, L. K., & Roweis, S. T. "Think globally, fit locally: Unsupervised learning of low dimensional manifolds." Journal of Machine Learning Research, vol. 4, Jun. 2003, pp. 119-155.
