Principal Component Analysis (PCA)
Introduction
Principal Component Analysis (PCA) is a statistical procedure that uses an orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of linearly uncorrelated variables called principal components. The number of principal components is less than or equal to the number of original variables. This transformation is defined in such a way that the first principal component has the largest possible variance (that is, accounts for as much of the variability in the data as possible), and each succeeding component, in turn, has the highest variance possible under the constraint that it is orthogonal to the preceding components. The resulting vectors (principal components) are an uncorrelated orthogonal basis set. PCA is sensitive to the relative scaling of the original variables.
Background and Theory
PCA is founded on the method of eigenvalue decomposition of a data covariance (or correlation) matrix or singular value decomposition (SVD) of a data matrix, usually after mean centering (and normalizing or using Z-scores) the data matrix for each attribute. The PCA transformation can be considered as identifying the "axes" that maximize the variance of the data.
Mathematical Foundations
Given a matrix of dimensions representing observations of variables, PCA aims to find a transformation matrix that transforms into a new matrix of dimensions in which each row is an observation described in the principal components space.
-
Standardization: Often, the first step is to standardize , resulting in a matrix , where each column has zero mean and unit variance.
-
Covariance Matrix Computation: Compute the covariance matrix from .
-
Eigen Decomposition: Solve for the eigenvalues and eigenvectors of .
-
Select Principal Components: Sort the eigenvectors by decreasing eigenvalues and choose the first eigenvectors. This forms the transformation matrix .
-
Transformation: Transform using to obtain the principal components .
Dimensionality Reduction
By selecting a subset of the principal components, PCA achieves dimensionality reduction while preserving as much of the data's variation as possible.
Procedural Steps
- Data Preparation: Center and, if necessary, standardize the variables.
- Covariance Matrix Analysis: Compute the covariance or correlation matrix.
- Eigenvalue Decomposition: Perform eigenvalue decomposition on the covariance matrix.
- Principal Component Selection: Decide the number of principal components to retain.
- Data Projection: Project the data onto the space spanned by the selected principal components.
Implmentation
Parameters
n_components:int
Number of principal components
Examples
from luma.reduction.linear import PCA
from sklearn.datasets import load_iris
import matplotlib.pyplot as plt
import numpy as np
iris_df = load_iris()
X = iris_df.data
y = iris_df.target
model = PCA(n_components=2)
X_trans = model.fit_transform(X)
fig = plt.figure(figsize=(11, 5))
ax1 = fig.add_subplot(1, 2, 1, projection='3d')
ax2 = fig.add_subplot(1, 2, 2)
for cl, m in zip(np.unique(y), ['s', 'o', 'D']):
X_cl = X[y == cl]
sc = ax1.scatter(X_cl[:, 0], X_cl[:, 1], X_cl[:, 2],
c=X_cl[:, 3],
marker=m,
label=iris_df.target_names[cl])
ax1.set_xlabel(iris_df.feature_names[0])
ax1.set_ylabel(iris_df.feature_names[1])
ax1.set_zlabel(iris_df.feature_names[2])
ax1.set_title('Original Iris Dataset')
ax1.legend()
cbar = ax1.figure.colorbar(sc, fraction=0.04)
cbar.set_label(iris_df.feature_names[3])
for cl, m in zip(np.unique(y), ['s', 'o', 'D']):
X_tr_cl = X_trans[y == cl]
ax2.scatter(X_tr_cl[:, 0], X_tr_cl[:, 1],
marker=m,
edgecolors='black',
label=iris_df.target_names[cl])
ax2.set_xlabel(r'$PC_1$')
ax2.set_ylabel(r'$PC_2$')
ax2.set_title(f'Iris Dataset after {type(model).__name__}')
ax2.legend()
plt.tight_layout()
plt.show()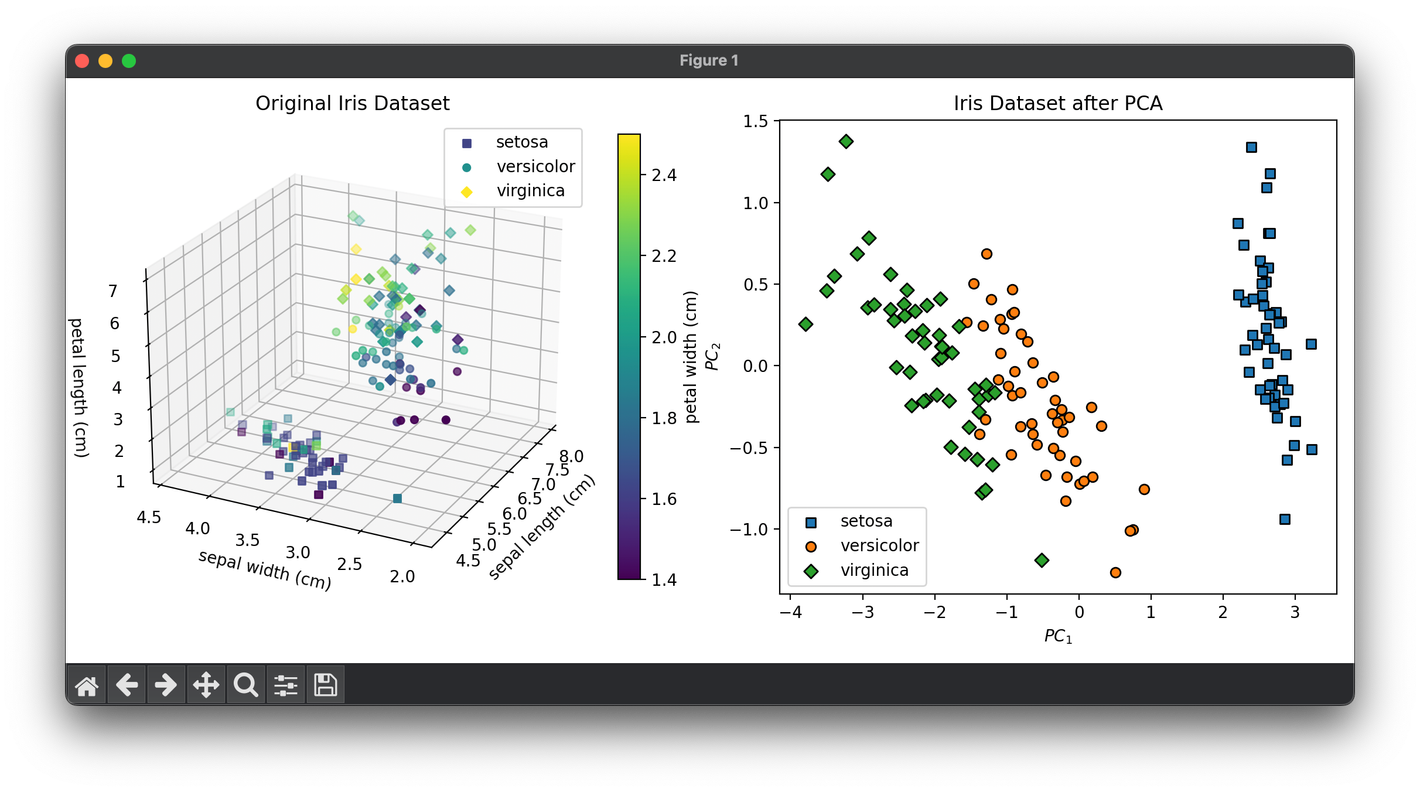
Applications
- Data Visualization: Reducing the dimensionality for visualization purposes.
- Noise Reduction: Eliminating noise by selecting a subset of principal components.
- Feature Extraction and Data Compression: Reducing the number of variables while retaining the essential information.
- Preprocessing for Machine Learning: Improving the performance of machine learning algorithms by reducing the dimensionality of the input data.
Strengths and Limitations
Strengths
- Versatility: Applicable to almost any data set, providing insights into the structure of the data.
- Data Compression: Efficiently reduces dimensionality, often with minimal loss of information.
- Simplicity: The algorithm is straightforward to implement and interpret.
Limitations
- Linearity: Assumes linear relationships among variables.
- Sensitivity to Scaling: Results depend on the scaling of the variables.
- Variance-based Selection: Principal components are selected based on variance, which might not always be the most relevant criterion.
Advanced Topics
- Kernel PCA: Extension of PCA for nonlinear dimensionality reduction through the use of kernels.
- Sparse PCA: Variation of PCA where some components are constrained to be zero for interpretability and efficiency.
- Incremental PCA: Algorithms designed for large datasets, allowing for PCA to be applied in a piecewise manner.
References
- Jolliffe, I. T. "Principal Component Analysis." Springer Series in Statistics. Springer, New York, NY, 2002.
- Abdi, H., and Williams, L.J. "Principal component analysis." Wiley Interdisciplinary Reviews: Computational Statistics, 2(4):433-459, 2010.
- Pearson, K. "On Lines and Planes of Closest Fit to Systems of Points in Space." Philosophical Magazine, 2(11):559–572, 1901.
- Hotelling, H. "Analysis of a complex of statistical variables into principal components." Journal of Educational Psychology, 24:417–441, 1933.
