Multinomial Mixture Model (MMM)
Introduction
Multinomial Mixture Models are probabilistic models used for clustering categorical data. These models assume that the data are generated from a mixture of several multinomial distributions, each representing a cluster. The model is particularly useful in applications where objects are represented by counts or frequencies of events, such as text document clustering, where documents are represented by word counts.
Background and Theory
In contrast to Gaussian Mixture Models (GMMs) that are suited for continuous data, Multinomial Mixture Models are designed for discrete data. The key assumption is that the observed data are generated from a finite mixture of multinomial distributions, with each distribution corresponding to a different underlying process or cluster.
Mathematical Formulation
Given a dataset of items, where each item is a -dimensional vector of counts, the probability of observing a particular item under a Multinomial Mixture Model is given by:
where:
- is the number of clusters (multinomial distributions) in the mixture.
- is the mixing coefficient for cluster , indicating the probability that an item belongs to cluster , with .
- is the probability of item under the multinomial distribution for cluster , characterized by the parameter vector , where is the probability of the -th category in the -th cluster, and .
Procedural Steps
Initialization
Choose the number of clusters . Initialize the mixing coefficients and the probability vectors for each cluster.
Expectation (E) Step
Calculate the posterior probabilities (responsibilities) that each item belongs to each cluster, given the current parameters. For item and cluster , this is:
Maximization (M) Step
Update the parameters and to maximize the expected log-likelihood of the observed data, given the current responsibilities:
- Update :
- Update : For each category in cluster , update :
Iteration and Convergence
Repeat the E and M steps until the change in the log-likelihood or the parameters between iterations falls below a predefined threshold.
Implementation
Parameters
n_clusters:int
Number of clusters(mixtures) to estimate
max_iter:int, default = 100
Maximum amount of iteration
tol:float, default = 0.00001
Tolerance for early convergence
Examples
Test on synthesized 2D multinomial dataset with 2 mixtures:
from luma.clustering.mixture import MultinomialMixture
from luma.visual.evaluation import ConfusionMatrix
import matplotlib.pyplot as plt
import numpy as np
def generate_multi_dataset(n_samples: int,
component_probs: list,
mixture_weights: list) -> tuple:
n_components = len(component_probs)
dataset, labels = [], []
for _ in range(n_samples):
component = np.random.choice(range(n_components), p=mixture_weights)
sample = np.random.multinomial(1, component_probs[component])
dataset.append(sample)
labels.append(component)
return np.array(dataset), np.array(labels)
X, y = generate_multi_dataset(n_samples=300,
component_probs=[[0.2, 0.8],
[0.7, 0.3]],
mixture_weights=[0.5, 0.5])
mmm = MultinomialMixture(n_clusters=2, max_iter=1000)
mmm.fit(X)
fig = plt.figure(figsize=(10, 5))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
n_clusters = 2
bincounts = [np.bincount(X[y == i, 0]) for i in range(2)]
width = 0.2
ax1.bar(np.arange(n_clusters) - width / 2, bincounts[0],
width=width,
label='Cluster 0')
ax1.bar(np.arange(n_clusters) + width / 2, bincounts[1],
width=width,
label='Cluster 1')
ax1.set_xticks([0, 1])
ax1.set_ylabel('Conut')
ax1.set_title('Frequency Counts')
ax1.legend()
conf = ConfusionMatrix(y_true=y, y_pred=mmm.labels)
conf.plot(ax=ax2, show=True)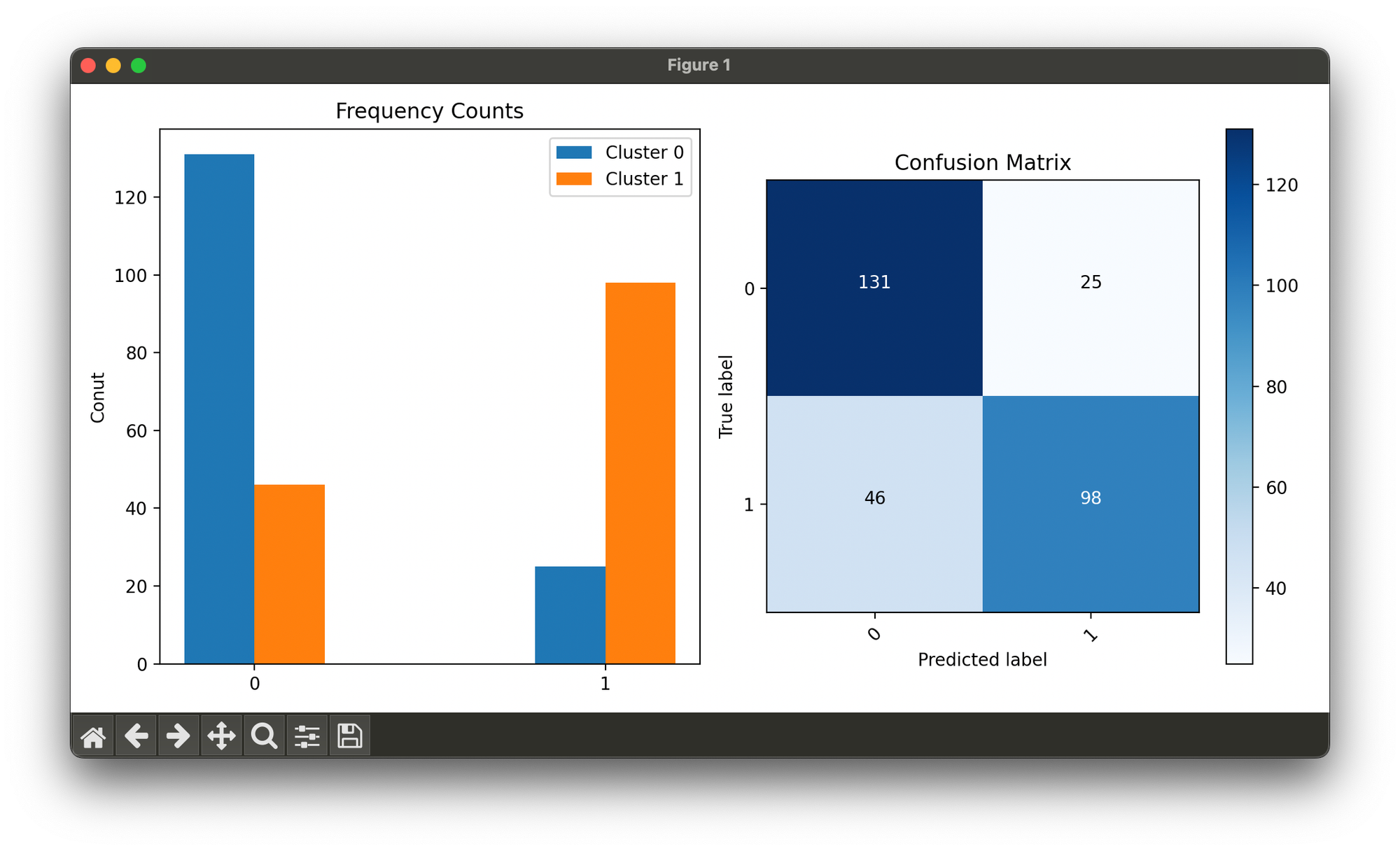
Applications
- Text Clustering: Grouping documents into topics based on word frequencies.
- Market Basket Analysis: Clustering customer purchase patterns to identify common baskets of products.
- Genetic Sequence Analysis: Clustering sequences into groups based on the frequency of nucleotides or amino acids.
Strengths and Limitations
Strengths
- Suitable for Categorical Data: Specifically designed to handle discrete count data efficiently.
- Interpretable Clusters: The parameters of the multinomial distributions can provide insights into the characteristics of each cluster.
Limitations
- Assumption of Independence: Assumes that the features (e.g., words in text clustering) are conditionally independent given the cluster, which may not always be true.
- Selection of Number of Clusters: Like other mixture models, determining the optimal number of clusters requires additional criteria or validation methods.
Advanced Topics
- Model Selection: Techniques like Bayesian Information Criterion (BIC) or Cross-Validation can be used to select the number of clusters.
- Extension to Other Distributions: For different types of data, the multinomial distribution can be replaced with other appropriate distributions within the mixture model framework.
References
- McLachlan, Geoffrey, and Thriyambakam Krishnan. The EM algorithm and extensions. John Wiley & Sons, 2007.
- Blei, David M., Andrew Y. Ng, and Michael I. Jordan. "Latent dirichlet allocation." Journal of machine Learning research 3.Jan (2003): 993-1022.
