Tree-based Methods
- Here we describe
tree-basedmethods for regression and classification - These involve
stratifyingorsegmentingthe predictor space into a number of simple regions - Since the set of splitting rules used to segment the predictor space can be summarized in a tree, these types of approaches are known as
decision-treemethods
Pros and Cons
Tree-basedmethods are simple and useful forinterpretation- However they typically are not competitive with the best supervised learning approaches in terms of prediction accuracy
- Hence we also discuss
bagging,random forests, andboosting - Combining a large number of trees can often result in dramatic improvements in prediction accuracy, at the expense of some loss interpretation
The Basics of Decision Trees
- Decision trees can be applied to both
regreesionandclassificationproblems - We first consider
regressionproblems, and then move on toclassification
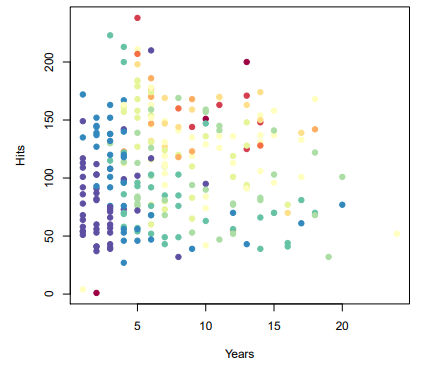
Baseball salary data : how would you stratify it?
- Salary is color-coded from low (blue, green) to high (yello, red)
 |  |
|---|
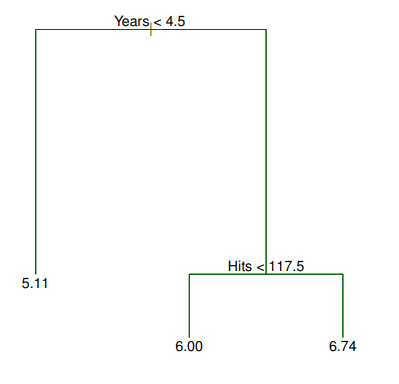
- The tree has two
internalnodes and threeterminalnodes, or leaves - The number in each leaf is the
meanof the response for the observations that fall there - Overall, the tree stratifies or segments the players into three regions of predictor space
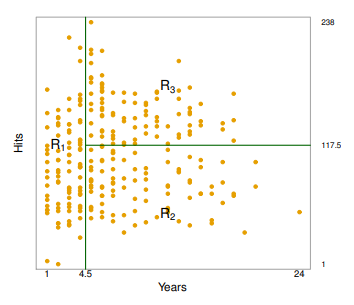
Terminology for Trees
- In keeping with the
treeanalogy, the regions , and are known asterminal nodes Decision treesare typically drawnupside down, in the sense that the leaves are at the bottom of the tere- The points along the tree where the predictor space is split are referred to as
internal nodes - In the hitters tree, the two
internal nodesare indicated by the text
and
Interpretation of Results
- is the most important factor in determining and players with less experience earn lower salaries than more experienced players
- Given that a player is less experienced, the number of that he made in the previous year seems to play little role in his
- But among players who have been in the major leagues for five or more years, the number of made in the previous year does affect , and players who made more last year tend to have higher salaries
- Surely an over-simplification, but compared to a
regressionmodel, it is easy to display,interpretand explain
Details of the tree-building process
- We divide the
predictor space- that is, the set of possible values for - into distinct and non-overlapping regions - For every observation that falls into the region , we make the same prediction which is simply the mean of the response values for the training observations in
- In theory, the regions could have any shape,. However, we choose to divide the predictor space into
high-dimensional rectangls, orboxes, for simplicity and for ease ofinterpretationof the resulting predictive model - The goal is to find boxes that
minimizestheRSS, given bywhere is the mean response for the training observations within the th box
- Unfortunately, it is computationally infeasible to consider every possible partition of the feature space info boxes
- For this reason, we take a
top-down,greedyapproach that is known asrecursive binary splitting - The approach is
top-downbecause it begins at the top of the tree and then successively splits thepredictor space - Each split is indicated via two new branches further down on the tree
- It is
greedybecause at each step of the tree-building process, thebestsplit is made at the particular step, rather than looking ahead and picking a split that will lead to a better tree in some future step
Greedy Approach of Tree-building Process
- We first select the predictor and the cutpoint such that splitting the predictor space into the regionsleads to the greatest possible reduction in
RSS - Next, we repeat the process, looking for the
best predictorandbest cutpointin order to split the data further so as tominimizetheRSSwithin each of the resulting regions - However, this time, instead of splitting the entire predictor space, we split one of the two previously identified regions
- Again, we look to split one of these regions further, so as to
minimizetheRSS. The process continues until a stopping criterion is reachedFor instance, we may continue until no region contains more than five observations
Predictions
- We predict the response for a given test observation using the
meanof the training observations in the region to which that test observation belongs - A five-region example of this approach is shown in the next
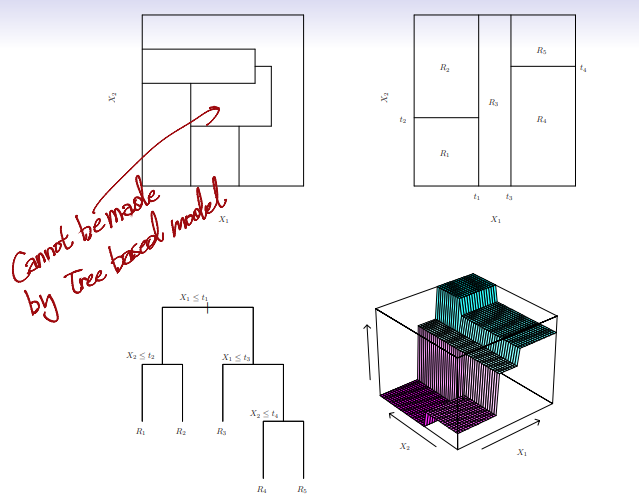
Top Left: A partition of two-dimensional feature space that could not result from recursive binary splittingTop Right: The output of recursive binary splitting on a two-dimensional exampleBottom Left: A tree corresponding to the partition in the top right pannelBottom Right: A perspective plot of the prediction surface corresponding to that tree
Pruning a tree
- The process described above may produce good predictions on the training set, but is likely to
overfitthe data, leading to poor test set perfomance - A
smaller treewith fewer splits (that is, fewer regions ) might lead tolower varianceandbetter interpretationat the cost oflittle bias - One possible alternative to the process described above is to
grow the treeonly so long as thedecrease in the RSSdue to each splitexceedssome (high)threshold - This strategy will result in
smaller trees, but is tooshort-sighteda seemingly worthless split early on in the tree might be followed by a very good split - that is, a split that leads to a large reduction in RSS later on
- A
better strategyis to grow avery large tree, and thenpruneit back in order to obtain asubtree Cost complexity pruning- also known asweakest link pruning- is used to do this- We consider a sequence of trees indexed by a
nonnegativetuning parameter - For each value of there corresponds a
subtreesuch thatRSS:- Number of
Terminal Node:
- Here indicates the number of
terminal nodesof the Tree is the rectangle(i.e. the subset of predictor space) corresponding to the thterminal node, and is the mean of the trainin observations in
Choosing the best subtree
- The tuning parameter controls a
trade-offbetween the subtree'scomplexityand itsfitto the training data - We select an
optimal valueusingcross-validation - We then return to the full data set and obtain the subtree corresponding to
Summary : tree algorithm
- Use
recursive binary splittingto grow a large tree on the training data,stoppingonly when eachterminal nodehasfewerthan someminimumnumber of observations - Apply cost
complexitypruning to the large tree in order to obtain a sequence ofbest subtrees, as a function of - Use -fold
cross-validationto choose . For each- Repeat Steps 1 and 2 on the th fraction of the training data, excluding the th fold
- Evaluate the
mean squared prediction erroron the data in the left-out th fold, as a function of
- Return the
subtreefromStep 2that corresponds to the chosen value of
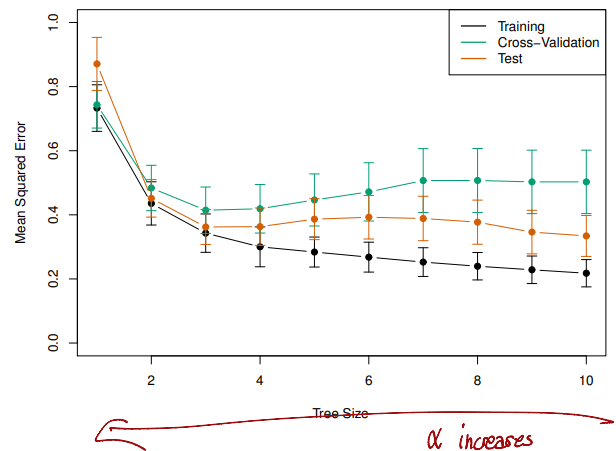
Then we could pick value when the
Tree Sizehas become 3 using 1 std rule
Classification Trees
- Very similar to a
regression tree, except that it is used to predict aqualitative responserather than aquantitativeone - For a
classification tree, we predict that each observation belongs to themost commonly occurring classof training observations in the region to which it belongs - Just as in the
regressionsetting, we userecursive binary splittingto grow aclassification tree - In the
classificationsetting,RSScannot be used as a criterion for for making thebinary splits
- A natural alternative to
RSSis theclassification error rateHere represents the proportion of training observations in the th region that are from the th class. - However
classification erroris not sufficiently sensitive fortree-growing, and in practice two other measures are preferable
Gini index and Deviance
- The
Gini indexis defined by - For this reason the
Gini indexis referred to as a measure of nodepurity- a small value indicates that anodecontains predominantly observations from a single class - An alternative to the
Gini indexiscross-entropy, given by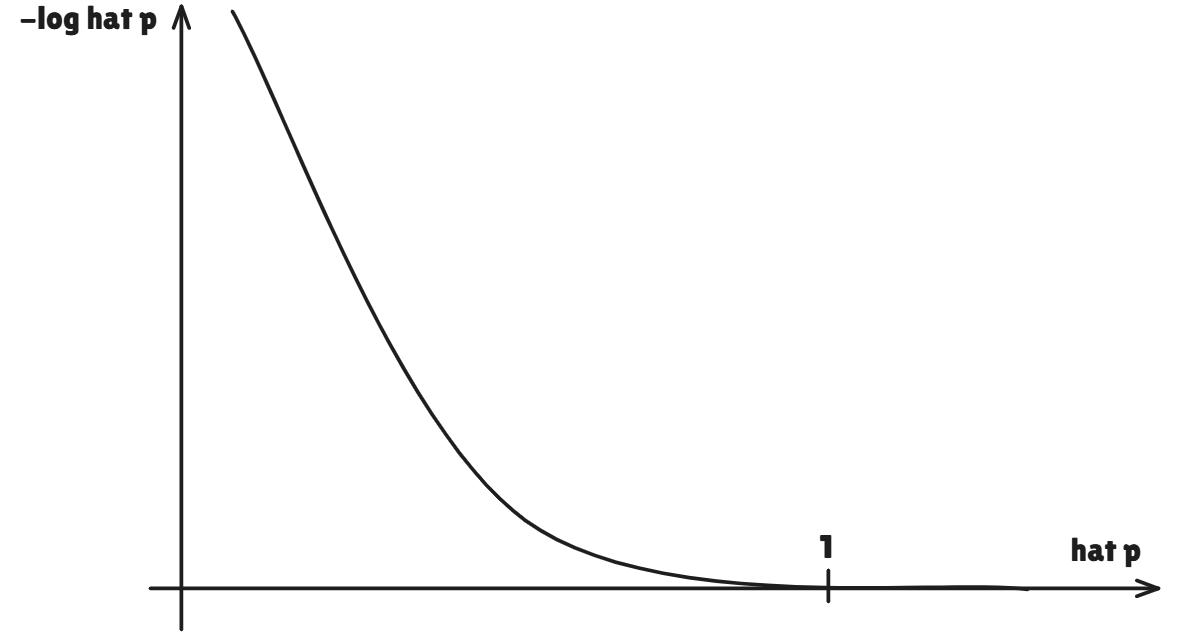
- It turns out that the
Gini indexand thecross-entropyare very similar numerically
Trees Versus Linear Models
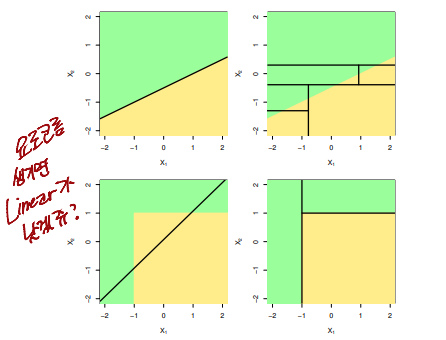
- Top Row : True
linearboundary - Bottom Row : true
non-linear boundary - Left Column :
Linear model - Right Column :
non-linear model
Advantages and Disadvantages of Trees
- Advantages
- Trees are very easy to explain to people. They are even easier to explain than
linear regression - Some people believe that
decision treesmore closely mirror humandecision-makingthan do theregressionandclassificationapproaches seen in previous chapters - Trees can be displayed
graphicallyand are easilyinterpretedeven by a non-expert - Trees can easily handle
qualitativepredictors without the need to create dummy variables
- Trees are very easy to explain to people. They are even easier to explain than
- Disadvantages
- Unfortunately, trees generally do not have the same level of
predictive accuracyas some of the otherregressionandclassificationapproaches seen in this book
- Unfortunately, trees generally do not have the same level of
Bagging
Bootstrap aggregation, orbagging, is a general-purpose procedure forreducing the varianceof a statistical learning method- Recall that given a set of independent observations , each with variance the
varianceof themeanof the observations is given by - In other words,
averaginga set of observationsreduces variance - Of course, this is not practical because we generally do not have access to multiple training sets
- Instead, we can
bootstrap, by taking repeated samples from the (single) training data set - In this approach we generate different
bootstrappedtraining data sets - We train our method on the th
bootstrappedtraining set in order to get , the prediction at a point - We then average all the predictions to obtain
This is called
bagging
Bagging classification trees
- The above prescription applied to
regression trees - For
classification trees: for each test observation, we record the class predicted by each of the trees, and take amajority vote
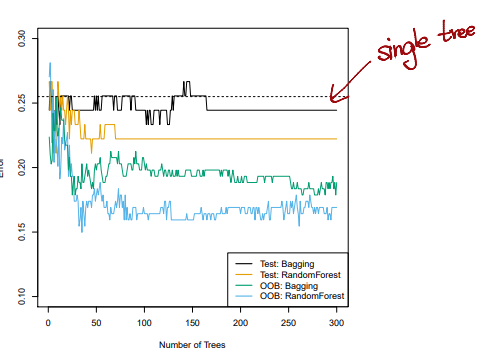
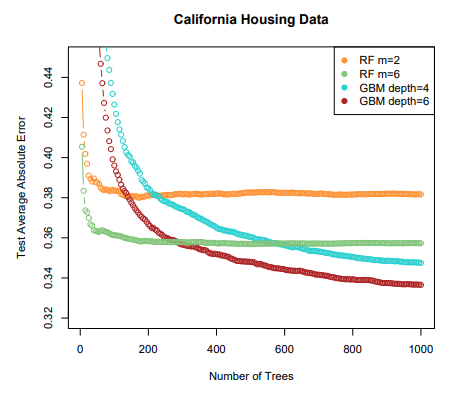
- The
test error(black and orange) is shown as a function of , the number ofbootstrappedtraining sets used Random forestswere applied with- The dashed line indicates the test error resulting from a
singleclassificaion tree - The green and blue traces show the
OOBerror, which in this case is considerably lower
- The
Out-of-Bag Error Estimation
- It turns out that there is a very straightforward way to estimate the test error of a
bagged model - Recall that the key to
baggingis thattreesare repeatedly fit tobootstrappedsubsets of the observations - One can show that on
average, eachbaggedtree makes use of aroundtwo-thirdsof the observations - The remaining
one-thirdof the observations not used tofita givenbagged treeand referred to as theout-of-bag(OOB) observations - We can predict the response for the th observation using each of the trees in which that observation was
OOB - This will yield aroung predictions for the th observation, which we
average - This estimate is essentially the
LOOcross-validation errorforbagging, if islarge
Random Forests
Random forestsprovide an improvement overbagged treesby way of a small tweak thatdecorrelatesthe trees- This reduces the
variancewhen weaveragethe trees - As in
bagging, we build a number ofdecision treesonbootstrappedtraining samples - But when building these
decision trees, each time a split in a tree is considered, arandom selectionof predictors is chosen as split candidates from the full set of predictors - The split is allowed to use only one of those predictors
- A fresh selection of predictors is taken at each split, and typically we choose
That is, the number ofpredictorsconsidered at each split is approximately equal to the square root of the total number of prdictors
Boosting
- Like
bagging,boostingis a general approach that can be applied to many statistical learning methods forregressionorclassification - We only discuss
boostingfor decision trees - Recall that
bagginginvolves creating multiple copies of the original training data set using thebootstrap,fittinga seperate decision tree to each copy, and then combining all of the trees in order to create asingle predictive model - Notably, each tree is built on a
bootstrapdata set, independent of the other trees Boostingworks in a similar way, except that the trees are grownsequentially
Boosting algorithm for regression trees
- Set and for all in the training set
- For , repeat :
- Fit a tree with splits
terminal nodesto the training data - Update by adding in a
shrunkenversion of the new tree : - Update the residuals
- Fit a tree with splits
- Output the
boosted model
What is the idea behind this procedure?
- Unlike fitting a single large decision tree to the data, which amounts to
fitting the data hardand potentially overfitting, theboostingapproach insteadlearns slowly - Given the current model, we fit a decision tree to the residual from the model
- We then add this new
decision treeinto thefitted functionin order to update the residuals - Each of these trees can be rather
small, with just a fewterminal nodes, determined by the parameter in the algorithm - By
fittingsmall trees to theresiduals, we slowly improve in areas where it does not perform well - The
shrinkage parameterslows the process down even further, allowing more and different shaped trees to attack the residuals
Boosting for classification
Boostingforclassificationis similar in spirit toboostingforregression, but is a bit morecomplex- We will not go into detail here, nor do we in the text book
- Students can learn about the details in Elements of Statistical Learning, chapter 10
Tuning parameters for boosting
- The
number of trees
Unlikebaggingandrandom forests,boostingcanoverfitif is too large, although this overfitting tends to occur slowly if at all. We usecross-validationto select - The
shrinkage parameter
A small positive number. This controls the rate at whichboostinglearns. Typical values are 0.01 or 0.001, and the right choice can depend on the problem.
Very small can require using a very large value of in order to achieve good performance - The
number of splits
In each tree, which controls thecomplexityof theboosted ensemble
Often works well, in which case each tree is astump, consisting of a sinple split and resulting in an additive model.
More generally is theinteraction depth, and controls the interaction order of theboosted model, since splits can involve at most variables
Variable importance measure
- For
bagged/RFregression trees, we record the total amount that theRSSis decreased due to splits over a givenpredictor, averaged over all trees - A large value indicates an
important predictor - Similarly, for
bagged/RFclassification trees, we add up the total amount that theGini indexisdecreasedby splits over a given predictor,averagedover all trees

