Mini-Batch K-Means Clustering
Introduction
Mini-Batch K-Means is a variant of the K-Means clustering algorithm that aims to reduce the computational cost by using small, random samples of the data (mini-batches) for each iteration of the algorithm, instead of the full dataset. This approach significantly speeds up the convergence time and is particularly effective for large datasets, making it a popular choice for scalable machine learning applications.
Background and Theory
K-Means is a widely used clustering algorithm that partitions a dataset into (K) clusters by minimizing the variance within each cluster. The Mini-Batch K-Means algorithm introduces an optimization by processing small subsets of the dataset in each iteration, which drastically reduces the amount of computation needed to converge to a solution.
Mathematical Foundations
The objective function of the K-Means algorithm is to minimize the sum of squared distances between each data point and the centroid of its assigned cluster. For a dataset with data points, the objective function is defined as:
where represents the set of points in cluster , is the centroid of cluster , and is the squared Euclidean distance between point and centroid .
Procedural Steps
- Initialization: Randomly select centroids from the dataset as the initial cluster centers.
- Mini-Batch Processing: At each iteration, randomly sample a mini-batch of data points from the dataset.
- Cluster Assignment: Assign each data point in the mini-batch to the nearest centroid.
- Centroid Update: Update the centroids of the clusters based on the newly assigned points from the mini-batch.
- Iteration: Repeat steps 2-4 until the centroids stabilize or a predefined number of iterations is reached.
Mathematical Formulations
Centroid Update
The centroid update in Mini-Batch K-Means is performed using an incremental update formula to account for the mini-batch processing. The new centroid after processing a mini-batch is calculated as:
where is the mean of the points in the mini-batch assigned to cluster , and is the learning rate, which controls the extent to which the centroids are updated. The learning rate may decrease over time to ensure convergence.
Implementation
Parameters
n_clusters:int
Number of clusters
batch_size:int, default = 100
Size of a single batch
max_iter:int, default = 100
Number of iteration
Examples
Test on reduced wine dataset and find optimal number of clusters via inertia elbow plot:
from luma.preprocessing.scaler import StandardScaler
from luma.reduction.linear import PCA
from luma.clustering.kmeans import MiniBatchKMeansClustering
from luma.metric.clustering import Inertia
from luma.visual.evaluation import ClusterPlot
from sklearn.datasets import load_wine
import matplotlib.pyplot as plt
import numpy as np
X, y = load_wine(return_X_y=True)
sc = StandardScaler()
X_std = sc.fit_transform(X)
pca = PCA(n_components=2)
X_pca = pca.fit_transform(X_std)
scores, labels, models = [], [], []
for i in range(2, 10):
km = MiniBatchKMeansClustering(n_clusters=i,
batch_size=100,
max_iter=100)
km.fit(X_pca)
scores.append(Inertia.score(X_pca, km.centroids))
labels.append(km.labels)
models.append(km)
def derivate_inertia(scores: list) -> list:
diff = [0.0]
for i in range(1, len(scores) - 1):
diff.append(scores[i - 1] - scores[i + 1] / 2)
diff.append(0.0)
return diff
d_scores = derivate_inertia(scores)
best_n = np.argmax(d_scores)
fig = plt.figure(figsize=(10, 5))
ax1 = fig.add_subplot(1, 2, 1)
ax2 = fig.add_subplot(1, 2, 2)
clst = ClusterPlot(models[best_n], X_pca, cmap='viridis')
clst.plot(ax=ax1)
ax2.plot(range(2, 10), scores, marker='o', label='Inertia')
ax2.plot(range(2, 10), d_scores, marker='o', label='Absolute Derivative')
ax2.set_title('Inertia Plot')
ax2.set_xlabel('Number of Clusters')
ax2.set_ylabel('Inertia')
ax2.legend()
ax2.grid()
plt.tight_layout()
plt.show()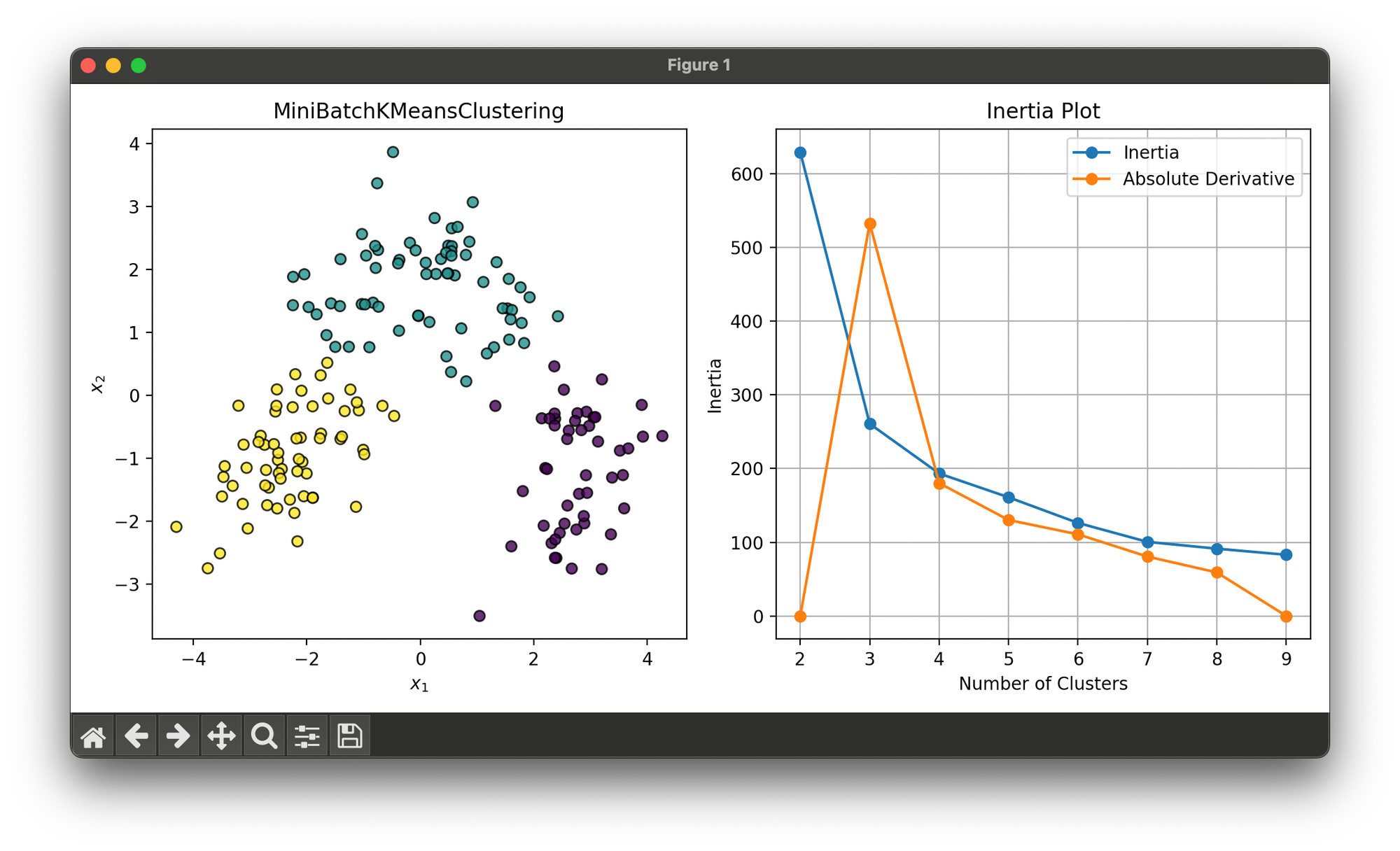
Applications
- Large-scale Data Clustering: Efficiently clustering large datasets that are infeasible to process with standard K-Means due to computational or memory constraints.
- Real-time Data Streaming: Clustering streaming data where the entire dataset is not available upfront and data arrives in increments.
- Image Quantization: Reducing the number of colors in an image for compression purposes without significantly affecting visual quality.
Strengths and Limitations
Strengths
- Scalability: Highly efficient for large datasets, reducing computational time and memory usage.
- Flexibility: Suitable for online clustering tasks where data arrives in a stream.
Limitations
- Approximation Quality: The centroids found by Mini-Batch K-Means may not be as accurate as those found by the standard K-Means due to the stochastic nature of the algorithm.
- Parameter Sensitivity: The choice of mini-batch size and learning rate can significantly affect the algorithm's performance and convergence.
Advanced Topics
- Optimal Mini-Batch Size: Investigating the impact of mini-batch size on the convergence rate and clustering quality.
- Learning Rate Strategies: Adaptive learning rate adjustments over iterations to improve convergence stability.
- Hybrid Approaches: Combining Mini-Batch K-Means with other clustering or dimensionality reduction techniques for enhanced performance in complex datasets.
References
- Sculley, David. "Web-scale k-means clustering." Proceedings of the 19th international conference on World wide web. 2010.
- Bradley, Paul S., Usama M. Fayyad, and Cory A. Reina. "Scaling clustering algorithms to large databases." Proceedings of the fourth international conference on knowledge discovery and data mining, AAAI Press, 1998.
